Abstract
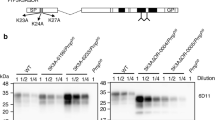
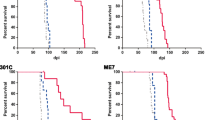
The N-terminal polybasic region of the normal prion protein, PrPC, which encompasses residues 23-31, is important for prion pathogenesis by affecting conversion of PrPC into the pathogenic isoform, PrPSc. We previously reported transgenic mice expressing PrP with residues 25-50 deleted in the PrP-null background, designated as Tg(PrP∆preOR)/Prnp 0/0 mice. Here, we produced two new lines of Tg(PrP∆preOR)/Prnp 0/0 mice, each expressing the mutant protein, PrP∆preOR, 1.1 and 1.6 times more than PrPC in wild-type mice, and subsequently intracerebrally inoculated RML and 22L prions into them. The lower expresser showed slightly reduced susceptibility to RML prions but not to 22L prions. The higher expresser exhibited enhanced susceptibility to both prions. No prion transmission barrier was created in Tg(PrP∆preOR)/Prnp 0/0 mice against full-length PrPSc. PrPSc∆preOR accumulated in the brains of infected Tg(PrP∆preOR)/Prnp 0/0 mice less than PrPSc in control wild-type mice, although lower in RML-infected Tg(PrP∆preOR)/Prnp 0/0 mice than in 22L-infected mice. Prion infectivity in infected Tg(PrP∆preOR)/Prnp 0/0 mice was also lower than that in wild-type mice. These results indicate that deletion of residues 25-50 only slightly affects prion susceptibility, the conversion of PrPC into PrPSc, and prion infectivity in a strain-specific way. PrP∆preOR retains residues 23-24 and lacks residues 25-31 in the polybasic region. It is thus conceivable that residues 23-24 rather than 25-31 are important for the polybasic region to support prion pathogenesis. However, other investigators have reported that residues 27-31 not 23-24 are important to support prion pathogenesis. Taken together, the polybasic region might support prion pathogenesis through multiple sites including residues 23-24 and 27-31.



Similar content being viewed by others
References
Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95:13363–13383
Stahl N, Borchelt DR, Hsiao K, Prusiner SB (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51:229–240
Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C (1993) Mice devoid of PrP are resistant to scrapie. Cell 73:1339–1347
Prusiner SB, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang SL, DeArmond SJ (1993) Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci USA 90:10608–10612
Manson JC, Clarke AR, McBride PA, McConnell I, Hope J (1994) PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration 3:331–340
Sakaguchi S, Katamine S, Shigematsu K, Nakatani A, Moriuchi R, Nishida N, Kurokawa K, Nakaoke R, Sato H, Jishage K et al (1995) Accumulation of proteinase K-resistant prion protein (PrP) is restricted by the expression level of normal PrP in mice inoculated with a mouse-adapted strain of the Creutzfeldt–Jakob disease agent. J Virol 69:7586–7592
Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C (1996) Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J 15:1255–1264
Turnbaugh JA, Unterberger U, Saa P, Massignan T, Fluharty BR, Bowman FP, Miller MB, Supattapone S, Biasini E, Harris DA (2012) The N-terminal, polybasic region of PrP(C) dictates the efficiency of prion propagation by binding to PrP(Sc). J Neurosci 32:8817–8830
Khalife M, Reine F, Paquet-Fifield S, Castille J, Herzog L, Vilotte M, Moudjou M, Moazami-Goudarzi K, Makhzami S, Passet B, Andreoletti O, Vilette D, Laude H, Beringue V, Vilotte JL (2016) Mutated but not deleted ovine PrP(C) N-terminal polybasic region strongly interferes with prion propagation in transgenic mice. J Virol 90:1638–1646
Yoshikawa D, Yamaguchi N, Ishibashi D, Yamanaka H, Okimura N, Yamaguchi Y, Mori T, Miyata H, Shigematsu K, Katamine S, Sakaguchi S (2008) Dominant-negative effects of the N-terminal half of prion protein on neurotoxicity of prion protein-like protein/doppel in mice. J Biol Chem 283:24202–24211
Scott MR, Kohler R, Foster D, Prusiner SB (1992) Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci 1:986–997
Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD (1985) Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA 82:4438–4442
Wilmut I, Hooper ML, Simons JP (1991) Genetic manipulation of mammals and its application in reproductive biology. J Reprod Fertil 92:245–279
Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C (1992) Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356:577–582
Yamaguchi N, Sakaguchi S, Shigematsu K, Okimura N, Katamine S (2004) Doppel-induced Purkinje cell death is stoichiometrically abrogated by prion protein. Biochem Biophys Res Commun 319:1247–1252
Reed J, Muench H (1938) A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497
Feraudet C, Morel N, Simon S, Volland H, Frobert Y, Creminon C, Vilette D, Lehmann S, Grassi J (2005) Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem 280:11247–11258
Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond SJ et al (1989) Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59:847–857
Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB (1998) Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 4:1157–1165
Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A (2002) Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 41:12868–12875
Safar JG, Geschwind MD, Deering C, Didorenko S, Sattavat M, Sanchez H, Serban A, Vey M, Baron H, Giles K, Miller BL, Dearmond SJ, Prusiner SB (2005) Diagnosis of human prion disease. Proc Natl Acad Sci USA 102:3501–3506
Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B (2005) The most infectious prion protein particles. Nature 437:257–261
Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR (2006) Isolation and characterization of a proteinase K-sensitive PrP(Sc) fraction. Biochemistry 45:15710–15717
Sajnani G, Silva CJ, Ramos A, Pastrana MA, Onisko BC, Erickson ML, Antaki EM, Dynin I, Vazquez-Fernandez E, Sigurdson CJ, Carter JM, Requena JR (2012) PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog 8:e1002547
Caughey B, Baron GS, Chesebro B, Jeffrey M (2009) Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem 78:177–204
Collinge J, Clarke AR (2007) A general model of prion strains and their pathogenicity. Science 318:930–936
Wadsworth JD, Asante EA, Collinge J (2010) Review: contribution of transgenic models to understanding human prion disease. Neuropathol Appl Neurobiol 36:576–597
Pan T, Wong BS, Liu T, Li R, Petersen RB, Sy MS (2002) Cell-surface prion protein interacts with glycosaminoglycans. Biochem J 368:81–90
Warner RG, Hundt C, Weiss S, Turnbull JE (2002) Identification of the heparan sulfate binding sites in the cellular prion protein. J Biol Chem 277:18421–18430
Taubner LM, Bienkiewicz EA, Copie V, Caughey B (2010) Structure of the flexible amino-terminal domain of prion protein bound to a sulfated glycan. J Mol Biol 395:475–490
Acknowledgements
We thank Dr. Stanley B. Prusiner for providing Zrch I Prnp 0/0 mice.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported in part by Pilot Research Support Program in Tokushima University, the Naito Foundation, JSPS KAKENHI grant numbers, JP26460557, JP26293212, MEXT KAKENHI grant numbers, J15H01560, and the Practical Research Project for Rare/Intractable Diseases of the Japan Agency for Medical Research and Development (AMED). H. M was partly supported by a Cooperative Research Grant of the Institute for Enzyme Research, Tokushima University.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors. All animal experiments in this study were performed according to the regulations and guidelines for animal ethics of The Guiding Principle for Animal Care and Experimentation of Tokushima University, the University of Occupational and Environmental Health, and Japanese Law for Animal Welfare and Care, with prior approval of The Ethics Committee of Animal Care and Experimentation of Tokushima University and the University of Occupational and Environmental Health (approval number T27-102, AE08-013).
Rights and permissions
About this article
Cite this article
Das, N.R., Miyata, H., Hara, H. et al. Effects of prion protein devoid of the N-terminal residues 25-50 on prion pathogenesis in mice. Arch Virol 162, 1867–1876 (2017). https://doi.org/10.1007/s00705-017-3295-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3295-3