Abstract
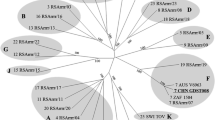
Phylogenetic networks and sequence analysis allow a more accurate understanding of the serotypes, genetic relationships and epidemiology of viruses. Based on gene sequences of the conserved segment 10 (NS3), bluetongue virus (BTV) can be divided into five topotypes. In this molecular epidemiology study, segment 10 sequence data of 11 isolates obtained from the Virology Section of the Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria, Onderstepoort, were analyzed and compared to sequence data of worldwide BTV strains available in the GenBank database. The consensus nucleotide sequences of NS3/A showed intermediate levels of variation, with the nucleotide sequence identity ranging from 79.72 % to 100 %. All 11 strains demonstrated conserved amino acid characteristics. Phylogenetic networks were used to identify BTV topotypes. The phylogeny obtained from the nucleotide sequence data of the NS3/A-encoding gene presented three major and two minor topotypes. The clustering of strains from different geographical areas into the same group indicated spatial spread of the segment 10 genes, either through gene reassortment or through the introduction of new strains from other geographical areas via trade. The effect of reassortment and genetic drift on BTV and the importance of correct serotyping to identify viral strains are highlighted.



Similar content being viewed by others
References
Spreull J (1905) Malarial catarrhal fever (bluetongue) of sheep in South Africa. J Comp Pathol Ther 18:321–337
Borden EC, Shoorijeh SJ, Murphy FA (1971) Physiochemical and morphological relationships of some athropod-borne viruses to bluetongue virus—a new taxonomic group. Physiochemical and serological studies. J Gen Virol 13:261–271
Davies FG, Mungai JN, Pini A (1992) A new bluetongue virus serotype isolated in Kenya. Vet Microbiol 31:25–32
Hofmann MA, Renzullo S, Mader M, Chaignat V, Worwa G et al (2008) Genetic characterization of toggenburg orbivirus, a new bluetongue virus, from goats, Switzerland. Emerg Infect Dis 14:1855–1861
Maan S, Maan NS, Nomikou K, Batten C, Antony F et al (2011) Novel bluetongue virus serotype from kuwait. Emerg Infect Dis 17:886–889
Maan S, Maan NS, Samuel AR, Rao SJ, Attoui H, Mertens PPC (2007) Analysis and phylogenetic comparisons of full-length VP2 genes of the 24 bluetongue virus serotypes. J Gen Virol 88:621–630
Du Toit RM (1944) The transmission of Bluetongue and horsesickness by Culicoides. Onderstepoort J Vet Sei Anim Indus 19:7–16
De Clercq K, De Leeuw I, Verheyden B, Vandemeulebroucke E, Vanbinst T et al (2008) Transplacental infection and apparently immunotolerance induced by a wild-type bluetongue virus serotype 8 natural infection. Transbound Emerg Dis 55:352–359
Desmecht D, Bergh RV, Sartelet A, Leclerc M, Mignot C et al (2008) Evidence for transplacental transmission of the current wild-type strain of bluetongue virus serotype 8 in cattle. Vet Rec 163:50–52
Vercauteren G, Miry C, Vandenbussche F, Ducatelle R, Van der Heyden S et al (2008) Bluetongue virus serotype 8-associated congenital hydranencephaly in calves. Transbound Emerg Dis 55:293–298
Menzies FD, McCullough SJ, McKeown IM, Forster JL, Jess SC et al (2008) Evidence for transplacental and contact transmission of bluetongue virus in cattle. Vet Rec 163:203–209
Miller PJ, Kim LM, Ip HS, Afonson CL (2009) Evolutionary dynamics of newcastle disease virus. Virology 391:64–72
He C, Xie Z, Han G, Dong J, Wang D, Liu J, Ma L, Tang X, Liu X, Pang Y et al (2009) Homologous recombination as an evolutionary force in the avian influenza A virus. Mol Biol Evol 26:177–187
Knowles NJ, Samuel AR (2003) Molecular epidemiology of foot-and-mouth disease virus. Virus Res 91:65–80
Holmes EC, Worobey M, Rambaut A (1999) Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol 16:405–409
Maan S, Maan NS, van Rijn PA, van Gennip RGP, Sanders A et al (2009) Full Genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains. PLoS One 5(4):e10323. doi:10.1371/journal.pone.0010323
Mellor PS, Carpenter S, Harrup L, Baylis M, Wilson A, Mertens PPC (2009) Bluetongue in Europe and the Mediterranean Basin. In: Mellor PS, Baylis M, Mertens PPC (eds) Bluetongue. Elsevier, London, pp 235–264
Vandenbussche F, De Leeuw I, Vandemeulebroucke E, De Clercq K (2009) Emergence of bluetongue serotypes in Europe, Part 1: description and validation of four real-time RT-PCR Assays for the serotyping of bluetongue viruses BTV-1, BTV-6, BTV-8 and BTV-11. Transbound Emerg Dis 56:346–354
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. In: Society for Molecular Biology and Evolution. Oxford University Press, pp 254–267
Maan S, Maan NS, Ross-Smith N, Batten CA, Shaw AE, Anthony SJ, Samuel AR, Darpel KE, Veronesi E, Oura CAL et al (2008) Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 377:308–318
Bonneau KR, Mullens BA, Maclachlan NJ (2001) Occurrence of genetic drift and founder effect during quasispecies evolution of the VP2 and NS3/NS3A genes of bluetongue virus upon passage between sheep, cattle, and Culicoides sonorensis. J Virol 75:8298–8305
Batten CA, Maan S, Shaw AE, Maan NS, Mertens PPC (2008) A European field strain of bluetongue virus derived from two parental vaccine strains by genome segment reassortment. Virus Res 137:56–63
Samal SK, el-Hussein A, Holbrook FR, Beaty BJ, Ramig RF (1987) Mixed infection of Culicoides variipennis with bluetongue virus serotypes 10 and 17: evidence for high frequency reassortment in the vector. J Gen Virol 68:2319–2329
Scott TW, Weaver SC, Mallampalli VL (1994) Evolution of mosquito-borne viruses. In: Morse SS (ed) Evolutionary biology of viruses. Raven Press, New York, pp 293–324
Novella IS, Hershey CL, Escarmis C, Domingo E, Holland JJ (1999) Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol 287:459–465
Weaver SC, Brault AC, Kang W, Holland JJ (1999) Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol 73:4316–4326
Wirblich C, Bhattacharya B, Roy P (2006) Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J Virol 80:460–473. doi:10.1128/JVI.80.1.460
Celma CCP, Roy P (2009) A viral non-structural protein regulates bluetongue virus trafficking and release. J Virol 83(13):6806–6816. doi:10.1128/JVI.00263-09
Gibbs EP, Greiner EC (1994) The epidemiology of bluetongue. Comp Immunol Microbiol 17:207–220
Balasuriya UBR, Nadler SA, Wilson WC, Pritchard LI, Smythe AB, Savini G, Monaco F, De Santis P, Zhang N, Tabachnick WJ et al (2008) The NS3 proteins of global strains of bluetongue virus evolve into regional topotypes through negative (purifying) selection. Vet Microbiol 126:91–100
Van Niekerk M, Freeman M, Paweska JT, Howell PG, Guthrie AJ, Potgieter AC, Van Staden V, Huisman H (2003) Variation in the NS3 gene and protein in South Africa isolates of bluetongue and equine encephalosis viruses. J Gen Virol 84:581–590
Huismans H, van Staden V, Fick WC, van Niekerk M, Meiring TL (2004) A comparison of different orbivirus proteins that could affect virulence and pathogenesis. Vet Ital Ser 40:417–425
Carpi G, Holmes EC, Kitchen A (2010) The evolutionary dynamics of bluetongue virus. J Mol Evol 70(6):583–592
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C content biases. Mol Biol Evol 9(4):678–687
Bryant D, Moulton V (2004) Neighbor-Net: an agglomerative algorithm for the construction of planar phylogenetic networks. Mol Biol Evol 2:255–265
Hwang GY, Yang YY, Chiou JF, Li JK (1992) Sequence conservation among the cognate non-structural NS3:3A protein genes of six bluetongue viruses. Virus Res 23:151–161
Pierce CM, Balasuriya UB, MacLachlan NJ (1998) Phylogenetic comparison of the S10 genes of field and laboratory strains of bluetongue virus from the United States. Virus Res 55:15–27
Ferrari G, De Liberato C, Scavia G, Lorenzetti R, Zini M, Farina F, Magliano A, Cardeti G, Scholl F, Guidoni M et al (2005) Active circulation of bluetongue vaccine virus serotype-2 among unvaccinated cattle in central Italy. Prev Vet Med 68(2–4):103–113
Barros SC, Ramos F, Luis TM, Vaz A, Duarte M, Henriques M, Cruz B, Fevereiro M (2007) Molecular epidemiology of bluetongue virus in Portugal during 2004–2006 outbreak. Vet Microbiol 124:25–34
Huismans H, Erasmus BJ (1981) Identification of the serotype-specific and group specific antigens of bluetongue virus. Onderstepoort J Vet 48:51–58
Mertens PPC (1999) Orbiviruses and coltiviruses—general features. In: Webster RG, Granoff A (eds) Encyclopaedia of virology, 2nd edn. Academic Press, London, pp 1043–1061
Listes E, Monaco F, Labrovic A, Paladini C, Leone A, Di GL, Camma C, Savini G (2009) First evidence of bluetongue virus serotype 16 in Croatia. Vet Microbiol 138:92–97
Gerdes GH (2004) A South African overview of the virus, vectors, surveillance and unique features of bluetongue. Vet Ital Ser 40(3):39–42
Delle-Porta AJ, Sellers RF, Herniman KA, Littlejohns IR, Cybinski DH, St George TD, McPhee DA, Snowdon WA, Campbell J, Cargill C et al (1983) Serological studies of Australian and Papua New Guinean cattle and Australian sheep for the presence of antibodies against bluetongue group viruses. Vet Microbiol 8:147–162
St George TD, Cybinski DH, Della-Porta AJ, McPhee DA, Wark MC, Bainbridge MH (1980) The isolation of two bluetongue viruses from healthy cattle in Australia. Aust Vet J 56:562–563
Sendow I, Pritchard LI, Eaton BT, Daniels PW (1997) Genotypic relationships of bluetongue viruses from Indonesia. Arbovirus Res Aust 7:263–266
Pritchard LI, Sendow I, Lunt R, Hassan R, Kattenbelt J, Gould AR, Daniels PW, Eaton BT (2004) Genetic diversity of bluetongue viruses in south east Asia. Virus Res 101:193–201
Cunha RG (1990) Neutralizing antibodies for different serotypes of bluetongue virus in sera of domestic ruminants from Brazil. Rev Bras Med Vet 12:3–7
Bonneau KR, MacLachlan NJ (2004) Genetic diversification of field strains of bluetongue virus. Vet Ital Ser 40:446–447
Bonneau KR, Zhang N, Zhu J, Zhang F, Li Z et al (1999) Sequence comparison of the L2 and S10 genes of bluetongue viruses from the United States and the People’s Republic of China. Virus Res 61:153–160
Gould AR, Pritchard LI (1990) Relationships amongst bluetongue viruses revealed by comparison of capsid and outer coat protein nucleotide sequences. Virus Res 17:31–52
Singh KP, Maan S, Samuel AR, Rao S, Meyer A, Mertens PPC (2004) Phylogenetic analysis of bluetongue virus genome segment 6 (encoding VP5) from different serotypes. Vet. Ital 40:479–483
Maan S, Maan NS, Nomikou K, Veronesi E, Bachanek-Bankowska K et al (2011) Complete Genome Characterisation of a Novel 26th Bluetongue Virus Serotype from Kuwait. Plos ONE. doi:10.1371/journal.pone.0026147
Acknowledgments
We thank the University of Pretoria, Department Veterinary Tropical Diseases, for giving us the opportunity to do the work. Dr. Melvyn Quan, Mr. Louwrens Snyman, Dr. Peter Coetzee and Miss Karen Eberhson are acknowledged for their assistance, and Dr. Otto Koekemoer, for information on sequences. Funding was received from the National Research Foundation and the Meat Industry Trust.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This study was approved by the Research Committee (V079/12).
Conflict of interest
The authors declare that they have no financial or personal relationships which may have inappropriately influenced the outcomes and conclusions they may have drawn.
Additional information
Nucleotide sequences are available in the GenBank database under the following accession numbers: BTV-2 is KJ506696, BTV-7 is KJ506695, BTV-8 is KJ506703, BTV-11 is KJ506694, BTV-16 is KJ506697, BTV-19 is KJ506699, BTV-20 is KJ506702, BTV-21 is KJ506701, BTV-22 is KJ506698, BTV-22 is KJ506700, BTV-S299 is KJ506704.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Steyn, J., Venter, E.H. Sequence analysis and evaluation of the NS3/A gene region of bluetongue virus isolates from South Africa. Arch Virol 161, 947–957 (2016). https://doi.org/10.1007/s00705-015-2741-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2741-3