Abstract
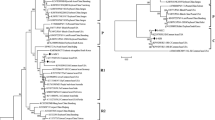
Molecular variability was assessed for 18 isolates of cowpea mild mottle virus (CPMMV, genus Carlavirus, family Betaflexiviridae) found infecting soybean in various Brazilian states (Bahia, Goiás, Maranhão, Mato Grosso, Minas Gerais, Pará) in 2001 and 2010. A variety of symptoms was expressed in soybean cv. CD206, ranging from mild (crinkle/blistering leaves, mosaic and vein clearing) to severe (bud blight, dwarfing, leaf and stem necrosis). Recombination analysis revealed only one CPMMV isolate to be recombinant. Pairwise comparisons and phylogenetic analysis were performed for partial genomes (ORF 2 to the 3’ terminus) and for each ORF individually (ORFs 2 to 6), showing the isolates to be distinct. The topology of the phylogenetic tree could be related to symptoms, but not to the year of collection or geographical origin. Additionally, the phylogenetic analysis supported the existence of two distinct strains of the virus, designated CPMMV-BR1 and CPMMV-BR2, with molecular variations between them.



Similar content being viewed by others
References
Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba S, Pearson MN, Ryu KH, Saldarelli P, Yoshikawa N (2012) Family Betaflexiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, San Diego, pp 920–941
Almeida AMR, Piuga FF, Kitajima EW, Gaspar JO, Valentin N, Benato LC, Marin SRR, Bineck E, Belintani P, Nunes Junior J, Hoffmann L, Meyer MC (2003) Necrose da haste da soja. Série Documentos 221:1–48
Almeida AMR, Piuga FF, Marin SRR, Kitajima EW, Gaspar JO, de Oliveira TG, de Moraes TG (2005) Detection and partial characterization of a carlavirus causing stem necrosis of soybean in Brazil. Fitopatol Bras 30:191–194
Almeida AMR (2008) Viroses da soja no Brasil: sintomas, etiologia, controle. Série Documentos 306:1–62
Boni MF, Posada D, Feldman MW (2007) An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047
Brito M, Fernandez-Rodriguez T, Garrido MJ, Mejias A, Romano M, Marys E (2012) First report of Cowpea mild mottle Carlavirus on yardlong bean (Vigna unguiculata subsp. sesquipedalis) in Venezuela. Viruses 4:3804–3811
Brown JK, Frohlich DR, Rosell RC (1995) The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annu Rev Entomol 40:511–534
Brunt AA, Kenten RH (1973) Cowpea mild mottle, a newly recognized virus infecting cowpeas (Vigna unguiculata) in Ghana. Ann Appl Biol 74:67–74
Byrne DN (1999) Migration and dispersal by the sweet potato whitefly, Bemisia tabaci. Agric For Meteorol 97:309–316
Carvalho SL, Silva FN, Zanardo LG, Almeida AMR, Zerbini FM, Carvalho CM (2013) Production of polyclonal antiserum against Cowpea mild mottle virus coat protein and its application in virus detection. Trop Plant Pathol 38:49–54
Chare ER, Holmes EC (2004) Selection pressures in the capsid genes of plant RNA viruses reflect mode of transmission. J Gen Virol 85:3149–3157
Chare ER, Holmes EC (2006) A phylogenetic survey of recombination frequency in plant RNA viruses. Arch Virol 151:933–946
Clark MF, Lister RM, Bar-Joseph M, Arthur Weissbach HW (1986) ELISA techniques. Methods enzymol. Academic Press, London, pp 742–766
Costa AS, Gaspar JO, Vega J (1983) Mosaico angular do feijão jalo causado por um carlavírus transmitido pela mosca branca Bemisia tabaci. Fitopatol Bras 8:325–327
De Souza J, Fuentes S, Savenkov EI, Cuellar W, Kreuze JF (2013) The complete nucleotide sequence of sweet potato C6 virus: a carlavirus lacking a cysteine-rich protein. Arch Virol 158(6):1393–1396
Dellaporta SL, Woud J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Report 1:19–21
Domingo E, Holland JJ (1997) RNA virus mutations and fitness for survival. Annu Rev Microbiol 51:151–178
Duarte PSG, Galvino-Costa SB, Ribeiro SRP, Figueira AR (2012) Complete genome sequence of the first Andean strain of Potato virus S from Brazil and evidence of recombination between PVS strains. Arch Virol 157:1357–1364
Duffy S, Shackelton LA, Holmes EC (2008) Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276
Edgar R (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BioMed Cent Bioinforma 5:113
Fontes FVHM, Colombo CA, Lourenção AL (2010) Caracterização molecular e divergência genética de Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae) em diferentes culturas e locais de cultivo. Neotrop Entomol 39(2):221–226
Frohlich DR, Torres-Jerez I, Bedford ID, Markham PG, Brown JK (1999) A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol 8:1683–1691
Garcia-Arenal F, Fraile A, Malpica JM (2001) Variability and genetic structure of plant virus populations. Annu Rev Phytopathol 39:157–186
Garcia-Arenal F, Fraile A, Malpica JM (2003) Variation and evolution of plant virus populations. Int Microbiol 6:225–232
Gaspar JO, Belintani P, Almeida AM, Kitajima EW (2008) A degenerate primer allows amplification of part of the 3′-terminus of three distinct carlavirus species. J Virol Methods 148:283–285
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582
Holmes EC (2009) The evolutionary genetics of emerging viruses. Annu Rev Ecol Evol Sist 40:353–372
Iwaki M, Thongmeearkon P, Prommin M, Honda Y, Hibi J (1982) Whitefly transmission and some properties of Cowpea mild mottle virus on soybean in Thailand. Plant Dis 66:265–268
Jeyanandarajah P, Brunt AA (1993) The natural occurrence, transmission, properties and possible affinities of Cowpea mild mottle virus. J Phytopathol 137:148–156
Laguna IG, Arneodo JD, Rodríguez-Pardina P, Fiorona M (2006) Cowpea mild mottle virus infecting soybean crops in northwestern Argentina. Fitopatol Bras 31:317
Lima AT, Sobrinho RR, Gonzalez-Aguilera J, Rocha CS, Silva SJ, Xavier CA, Silva FN, Duffy S, Zerbini FM (2013) Synonymous site variation due to recombination explains higher genetic variability in begomovirus populations infecting non-cultivated hosts. J Gen Virol 94:418–431
Lima LHC, Campos L, Moretzsohn MC, Návia D, Oliveira MRV (2002) Genetic diversity of Bemisia tabaci (Genn.) populations in Brazil revealed by RAPD markers. Genet Mol Biol 25:217–223
Martelli GP, Adams MJ, Kreuze JF, Dolja VV (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45:73–100
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563
Martin DP, Posada D, Crandall KA, Williamson C (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrovir 21:98–102
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Marubayashi JM, Yuki VA, Wutke EB (2010) Transmissão do Cowpea mild mottle virus pela mosca branca Bemisia tabaci biótipo B para plantas de feijão e soja. Summa Phytopathol 36:158–160
Menzel W, Winter S, Vetten H (2010) Complete nucleotide sequence of the type isolate of Cowpea mild mottle virus from Ghana. Arch Virol 155:2069–2073
Milgroom MG, Peever TL (2003) Populations biology of plant pathogens. Plant Dis 87:608–617
Munyappa V, Reddy DVR (1983) Transmission of Cowpea mild mottle virus by Bemisia tabaci in a nonpersistent manner. Plant Dis 67:391–393
Naidu RA, Gowda S, Satyanarayana T, Boyko V, Reddy AS, Dawson WO, Reddy DV (1998) Evidence that whitefly-transmitted Cowpea mild mottle virus belongs to the genus Carlavirus. Arch Virol 143:769–780
Nicolaisen M, Nielsen SL (2001) Analysis of the triple gene block and coat protein sequences of two strains of Kalanchoe latent carlavirus. Virus Genes 22:265–270
Nylander JAA (2004) MrModeltest v2. Program distributed by the author
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new gemini viruses by frequent recombination. Virology 265:218–225
Pagan I, Del Carmen Cordoba-Selles M, Martinez-Priego L, Fraile A, Malpica JM, Jorda C, Garcia-Arenal F (2006) Genetic structure of the population of Pepino mosaic virus infecting tomato crops in Spain. Phytopathology 96:274–279
Perring TM (2001) The Bemisia tabaci species complex. Crop Prot 20:725–737
Posada D, Crandall KA (2001) Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA 98:13757–13762
Rojas MR, Gilbertson RL, Russel DR, Maxwell DP (1993) Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis 77:340–347
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Simon-Loriere E, Holmes EC (2011) Why do RNA viruses recombine? Nat Rev Microbiol 9:617–626
Singh A, Mahinghara B, Hallan V, Ram R, Zaidi A (2008) Recombination and phylogeographical analysis of Lily symptomless virus. Virus Genes 36:421–427
Singh L, Hallan V, Martin D, Ram R, Zaidi A (2012) Genomic sequence analysis of four new Chrysanthemum virus B isolates: evidence of RNA recombination. Arch Virol 157:531–537
Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34:126–129
Sztuba-Solinska J, Urbanowicz A, Figlerowicz M, Bujarski JJ (2011) RNA-RNA recombination in plant virus replication and evolution. Annu Rev Phytophathol 49:415–443
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tavasoli M, Shahraeen N, Ghorbani S (2009) Serological and RT-PCR detection of Cowpea mild mottle Carlavirus infecting soybean. J Gen Mol Virol 1:7–11
Zanardo LG, Silva FN, Bicalho AAC, Urquiza GPC, Lima ATM, Almeida AMR, Zerbini FM, Carvalho CM (2013) molecular and biological characterization of Cowpea mild mottle virus isolates infecting soybean in Brazil and evidence of recombination. Plant Pathol
Acknowledgements
This work was funded by FAPEMIG (APQ-0992/09), CNPq (474112/2008-0) and Funarbe grants to CMC. LGZ was supported by a scholarship from CNPq. We are grateful to two anonymous referees and the editor Dr. Stenger for suggestions that have considerably improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2013_1879_MOESM1_ESM.tif
Two-dimensional plot representing the percent sequence identities between the 18 Brazilian cowpea mild mottle virus (CPMMV) isolates for five open reading frames or ORFs (ORF 2, triple block genes [TGB 1]; ORF 3 [TGB 2]; ORF 4 [TGB 3]; ORF 5, coat protein [CP] and ORF 6, nucleic acid binding protein [NABP]) evaluated. Nucleotide sequence identities are above the diagonal, and amino acid sequence identities below. (TIFF 3061 kb)
Rights and permissions
About this article
Cite this article
Zanardo, L.G., Silva, F.N., Lima, A.T.M. et al. Molecular variability of cowpea mild mottle virus infecting soybean in Brazil. Arch Virol 159, 727–737 (2014). https://doi.org/10.1007/s00705-013-1879-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1879-0