Abstract
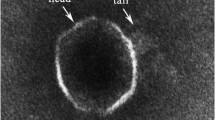
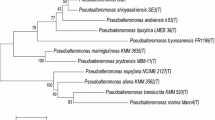
For molecular study of marine bacteria Pseudoalteromonas phenolica using bacteriophage, a novel bacteriophage, TW1, belonging to the family Siphoviridae, was isolated, and its genome was completely sequenced and analyzed. The phage TW1 genome consists of 39,940-bp-length double-stranded DNA with a GC content of 40.19 %, and it was predicted to have 62 open reading frames (ORFs), which were classified into functional groups, including phage structure, packaging, DNA metabolism, regulation, and additional function. The phage life style prediction using PHACTS showed that it may be a temperate phage. However, genes related to lysogeny and host lysis were not detected in the phage TW1 genome, indicating that annotation information about P. phenolica phages in the genome databases may not be sufficient for the functional prediction of their encoded proteins. This is the first report of a P. phenolica-infecting phage, and this phage genome study will provide useful information for further molecular research on P. phenolica and its phage, as well as their interactions.

Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA (2008) Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Egan S, Holmström C, Kjelleberg S (2001) Pseudoalteromonas ulvae sp. nov., a bacterium with antifouling activities isolated from the surface of a marine alga. Int J Syst Evol Microbiol 51:1499–1504
Gauthier G, Gauthier M, Christen R (1995) Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol 45:755–761
Holmström C, Kjelleberg S (1999) Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol 30:285–293
Holmström C, James S, Neilan BA, White DC, Kjelleberg S (1998) Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int J Syst Bacteriol 48:1205–1212
Isnansetyo A, Kamei Y (2003) Pseudoalteromonas phenolica sp. nov., a novel marine bacterium that produces phenolic anti-methicillin-resistant Staphylococcus aureus substances. Int J Syst Evol Microbiol 53:583–588
Ivanova EP, Kiprianova EA, Mikhailov VV, Levanova GF, Garagulya AD, Gorshkova NM, Vysotskii MV, Nicolau DV, Yumoto N, Taguchi T, Yoshikawa S (1998) Phenotypic diversity of Pseudoalteromonas citrea from different marine habitats and emendation of the description. Int J Syst Bacteriol 48:247–256
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
McNair K, Bailey BA, Edwards RA (2012) PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28:614–618
Oh YH, Jung C, Lee J (2011) Isolation and characterization of a novel agarase-producing Pseudoalteromonas spp. bacterium from the guts of spiny turban shells. J Microbiol Biotechnol 21:818–821
Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR Jr, Hendrix RW, Hatfull GF (2003) Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Wilcox SA, Toder R, Foster JW (1996) Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res 4:397–398
Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Acknowledgments
This work was supported in part by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2011-0012369), and the BK21 project of the Korean Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
H. Shin and J.-H. Lee contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shin, H., Lee, JH., Ahn, C.S. et al. Complete genome sequence of marine bacterium Pseudoalteromonas phenolica bacteriophage TW1. Arch Virol 159, 159–162 (2014). https://doi.org/10.1007/s00705-013-1776-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1776-6