Abstract
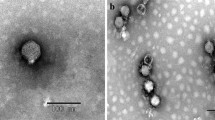
During sequencing of the genome of Clostridium perfringens strain 5147-97, a putative prophage was identified, located within a gene for a proposed flavodoxin oxidoreductase. Mitomycin C induction of this strain released a bacteriophage whose morphological features examined by electron microscopy indicated it belonged to the family Siphoviridae. This phage was hence designated as vB_CpeS-CP51. The 39,108-bp genome includes 50 predicted open reading frames (ORFs), including two that may affect sporulation, and two predicted tRNAs. To determine the ends of the prophage, PCR was performed using primers facing outwards from the proposed end genes. This confirmed the presence of a circularised genome in PEG-precipitated bacteriophage particles.


Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Bailly-Bechet M, Vergassola M, Rocha E (2007) Causes for the intriguing presence of tRNAs in phages. Genome Res 17:1486–1495
Goh S, Ong PF, Song KP, Riley TV, Chang BJ (2007) The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153:676–685
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Kim KP, Born Y, Lurz R, Eichenseher F, Zimmer M, Loessner MJ, Klumpp J (2012) Inducible Clostridium perfringens bacteriophages PhiS9 and PhiS63: Different genome structures and a fully functional sigK intervening element. Bacteriophage 2:89–97
Lindstrom M, Heikinheimo A, Lahti P, Korkeala H (2011) Novel insights into the epidemiology of Clostridium perfringens type A food poisoning. Food Microbiol 28:192–198
Morales CA, Oakley BB, Garrish JK, Siragusa GR, Ard MB, Seal BS (2012) Complete genome sequence of the podoviral bacteriophage PhiCP24R, which is virulent for Clostridium perfringens. Arch Virol 157:769–772
O’Flaherty S, Ross RP, Coffey A (2009) Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol Rev 33:801–819
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Lab. Press, Plainview
Schmitz JE, Ossiprandi MC, Rumah KR, Fischetti VA (2011) Lytic enzyme discovery through multigenomic sequence analysis in Clostridium perfringens. Appl Microbiol Biotechnol 89:1783–1795
Seal BS, Fouts DE, Simmons M, Garrish JK, Kuntz RL, Woolsey R, Schegg KM et al (2011) Clostridium perfringens bacteriophages PhiCP39O and PhiCP26F: genomic organization and proteomic analysis of the virions. Arch Virol 156:25–35
Sell TL, Schaberg DR, Fekety FR (1983) Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J Clin Microbiol 17:1148–1152
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123
Van Immerseel F, Rood JI, Moore RJ, Titball RW (2009) Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol 17:32–36
Volozhantsev NV, Oakley BB, Morales CA, Verevkin VV, Bannov VA, Krasilnikova VM, Popova AV et al (2012) Molecular characterization of podoviral bacteriophages virulent for Clostridium perfringens and their comparison with members of the Picovirinae. PLoS One 7:e38283
Volozhantsev NV, Verevkin VV, Bannov VA, Krasilnikova VM, Myakinina VP, Zhilenkov EL, Svetoch EA et al (2011) The genome sequence and proteome of bacteriophage PhiCPV1 virulent for Clostridium perfringens. Virus Res 155:433–439
Zimmer M, Scherer S, Loessner MJ (2002) Genomic analysis of Clostridium perfringens bacteriophage phi3626, which integrates into guaA and possibly affects sporulation. J Bacteriol 184:4359–4368
Acknowledgements
We thank Kathryn Cross (Imaging Platform, IFR) for microscopy, and K. I’Anson (IFR) and M. Brett for kind provision of strains. This research was funded by the Biotechnology and Biological Sciences Research Council (Grant BB/J004529/1), the EU-funded TORNADO grant (FP7-KBBE-222720) and the University of Messina.
Author information
Authors and Affiliations
Corresponding author
Additional information
The complete genome sequence of ΦCP51 was deposited in the GenBank database under accession number KC237729.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gervasi, T., Curto, R.L., Narbad, A. et al. Complete genome sequence of ΦCP51, a temperate bacteriophage of Clostridium perfringens . Arch Virol 158, 2015–2017 (2013). https://doi.org/10.1007/s00705-013-1647-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1647-1