Abstract
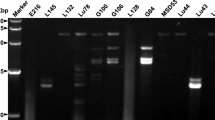
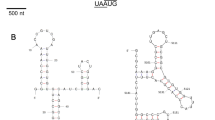
The Ninth Report of the International Committee on Taxonomy of Viruses (ICTV) reports only a few species whose members replicate in fungi. Most of these mycoviruses are described to replicate in phytopathogenic and commercially cultivated fungi. A few reports describe virus-like symptoms and virus-like particles in non-cultivated basidiocarps such as Boletus edulis, Laccaria spp. and Cantharellus spp. However, viral sequences from non-cultivated Agaricomycotina are not available yet. In this report, I present a partial sequence of a virus found in Clitocybe odora (Bull.:Fr.) P. Kumm var. odora coding for a putative RNA-dependent RNA polymerase (RdRp) and a small 20-kDa ORF that may encode a coat protein (CP). The sequence of the putative RdRp (ORF 1) of C. odora clusters with those of the Tanathephorus cucumeris virus RdRp and the Tuber aestivum mitovirus RdRp. In addition to sequence homology, Tanathephorus cucumeris virus shows a similar codon usage and TA content in the 5′- and 3′ non-translated regions, but it does not encode a putative CP. A viral DNA form proposed for Tanathephorus cucumeris virus was not found in Clitocybe odora. This viral sequence does not fit into any of the existing virus taxa.


Similar content being viewed by others
References
Blattný, C (1966) Viröse Mikrokefalie bei Laccaria sp. und weiteren Pilzarten. Česká Mykol 20:215
Blattný C, Pilát A (1957) Die Möglichkeit der Existenz von Virosen bei den Hutpilzen. Česká Mykol 11:205–211
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403
Huttinga H, Wichers HJ, Dielemann-van Zaayen A (1975) Filamentous and polyhedral virus-like particles in Boletus edulis. Neth J Pl Path 81:102–106
Knudsen H, Vesterholt J (eds) (2008) Funga Nordica. Copenhagen, Nordsvamp, p 968
Kreisel H (1983) Bildungsabweichungen an Fruchtkörpern (Teratologie). In: Henning M, Kreisel H (eds) Handbuch für Pilzfreunde, vol 5, 2nd edn. Gustav Fischer Verlag, Stuttgart, pp 26–68
Lakshman DK, Jian J, Tavantzis M (1998) A double-stranded RNA element from a hypovirulent strain of Rhizoctonia solani occurs in DNA form and is genetically related to the pentafunctional AROM protein of the shikimate pathway. Proc Natl Acad Sci USA 95:6425–6429
Permyakov E, Kretsinger R (2010) Calcium binding proteins. In: Uversky V (ed) Wiley Series in Protein and Peptide Science. Wiley, Hoboken, p 574
Potgieter AA, Page NA, Liebenberg J, Wright IM, Landt O, Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432
Stielow B, Menzel W (2010) Complete nucleotide sequence of TaV1, a novel totivirus isolated from a black truffle ascocarp (Tuber aestivum Vittad.). Arch Virol 155:2075–2078
Stielow B, Klenk HP, Winter S, Menzel W (2011) A novel Tuber aestivum mitovirus. Arch Virol 156:1107–1110
Stielow B, Bratek Z, Klenk HP, Winter S, Menzel W (2012) A novel mitovirus from the hypogeous ectomycorrhizal fungus Tuber excavatum. Arch Virol. doi:10.1007/s00705-012-1228-8
Tamura K, Peterson D, Stecher G, Nei M, Kumar S (2011) MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Ulbrich E (1926) Bildungsabweichungen bei Hutpilzen. Verhandl Bot Ver Prov Brandenburg 68:1–104
Verwoerd TC, Dekker BM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucl Acids Res 17:2362
Acknowledgments
I am grateful to Omar Darissa for dsRNA isolation, to Judith Mehrmann for electron microscopy, and to Peter Willingmann for help with sequence analysis and in preparing the manuscript. The picture from the symptomless basidiocarp was kindly provided by Georg Mueller (http://www.pilzepilze.de).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Heinze, C. A novel mycovirus from Clitocybe odora . Arch Virol 157, 1831–1834 (2012). https://doi.org/10.1007/s00705-012-1373-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1373-0