Abstract
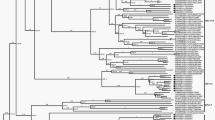
Sapporo virus belongs to the genus Sapovirus (family Caliciviridae) and has a non-segmented single-stranded, positive-sense RNA genome. This virus causes acute gastroenteritis in human, porcine and mink hosts. In this study, the complete genome of a Brazilian sapovirus isolate from a child with acute gastroenteritis was determined. A phylogenetic tree was constructed to analyze the genotype of this sapovirus (Sapo_BR-DF01), and possible intra- and inter-genogroups recombination events were evaluated in silico using the RDP3 program. Two inter-genogroup and two intra-genogroup recombination events were newly recognized in this study.



Similar content being viewed by others
References
Boni MF, Posada D, Feldman MW (2007) An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047
Chang KO, Sosnovtsev SS, Belliot G, Wang Q, Saif LJ, Green KY (2005) Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J Virol 79:1409–1416
Chiba S, Sakuma Y, Kogasaka R, Akihara M, Horino K, Nakao T, Fukui S (1979) Outbreak of gastroenteritis associated with calicivirus in an infant home. J Med Virol 4:249–254
Chiba S, Nakata S, Numata-Kinoshita K, Honma S (2000) Sapporo virus: history and recent findings. J Infect Dis 181(Suppl 2):S303–S308
Cunha JB, de Mendonca MCL, Miagostovich MP, Leite JPG (2010) Genetic diversity of porcine enteric caliciviruses in pigs raised in Rio de Janeiro State, Brazil. Arch Virol 155:1301–1305
Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz-Palacios GM, Pickering LK, Jiang X (2004) Genetic diversity among sapoviruses. Arch Virol 149:1309–1323
Fullerton SWB, Blaschke M, Coutard B, Gebhardt J, Gorbalenya A, Canard B, Tucker PA, Rohayem J (2007) Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J Virol 81:1858–1871
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582
Gutierrez-Escolano AL, Velazquez FR, Escobar-Herrera J, Saucedo CL, Torres J, Estrada-Garcia T (2010) Human caliciviruses detected in Mexican children admitted to hospital during 1998–2000, with severe acute gastroenteritis not due to other enteropathogens. J Med Virol :632–637
Guo M, Evermann JF, Saif LJ (2001) Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch Virol 146:479–493
Hansman GS, Natori K, Oka T, Ogawa S, Tanaka K, Nagata N, Ushijima H, Takeda N, Katayama K (2004) Cross-reactivity among sapovirus recombinant capsid proteins. Arch Virol 150:21–36
Hansman GS, Takeda N, Oka T, Oseto M, Hedlund KO, Katayama K (2005) Intergenogroup recombination in sapoviruses. Emerg Infect Dis 11:1916–1920
Hansman GS, Oka T, Sakon N, Takeda N (2007) Antigenic diversity of human sapoviruses. Emerg Infect Dis 13:1519–1525
Johnsen CK, Midgley S, Böttiger B (2009) Genetic diversity of sapovirus infections in Danish children 2005–2007. J Clin Virol 46:265–269
Katayama K, Miyoshi T, Uchino K, Oka T, Tanaka T, Takeda N, Hansman GS (2004) Novel recombinant sapovirus. Emerg Infect Dis 10:1874–1876
Kirkegaard K, Baltimore D (1986) The mechanism of RNA recombination in poliovirus. Cell 47:433–443
Kroneman A, Harris J, Vennema H, Duizer E, van Duynhoven Y, Gray J, Iturriza M, Bottiger B, Falkenhorst G, Johnsen C, von Bonsdorff CH, Maunula L, Kuusi M, Pothier P, Gallay A, Schreier E, Koch J, Szuecs G, Reuter G, Krisztalovics K, Lynch M, McKeown P, Foley B, Coughlan S, Ruggeri MF, Di Bartolo I, Vainio K, Isakbaeva E, Poljsak-Prijatelj M, Grom AH, Bosch A, Buesa J, Fauquier AS, Hernandez-Pezzi G, Hedlund KO, Koopmans M (2008) Data quality of 5 years of central norovirus outbreak reporting in the European Network for food-borne viruses. J Public Health 30:82–90
Lai MMC (1992) RNA Recombination in Animal and Plant Viruses. Microbiol Rev 56:61–79
Martella V, Lorusso E, Banyai K, Decaro N, Corrente M, Elia G, Cavalli A, Radogna A, Costantini V, Saif LJ, Lavazza A, DI Trani L, Buonavoglia C (2008) Identification of a porcine calicivirus related genetically to human sapoviruses. J Clin Microbiol 46:1907–1913
Martin DP (2009) Recombination detection and analysis using RDP3. Methods Mol Biol 537:185–205
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563
Martin D, Williamson C, Posada D (2005) RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262
Motomura K, Yokoyama M, Ode H, Nakamura H, Mori H, Kanda T, Oka T, Katayama K, Noda M, Tanaka T, Takeda N, Sato H. Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan. J Virol 84:8085–8097
Oka T, Katayama K, Ogawa S, Hansman GS, Kageyama T, Ushijima H, Miyamura T, Takeda N (2005) Proteolytic processing of sapovirus ORF1 polyprotein. J Virol 79:7283–7290
Oka T, Yamamoto M, Katayama K, Hansman GS, Ogawa S, Miyamura T, Takeda N (2006) Identification of the cleavage sites of sapovirus open reading frame 1 polyprotein. J Gen Virol 87:3329–3338
Padidam M, Sawer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265(2):218–225
Pang XLL, Lee BE, Tyrrell GJ, Preiksaitis JK (2009) Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004–2007. J Infect Dis 199:547–551
Phan TG, Khamrin P, Quang TD, Dey SK, Takanashi S, Okitsu S, Maneekarn N, Ushijima H (2007) Emergence of intragenotype recombinant sapovirus in Japan. Infect Genet Evol 7:542–546
Schlenker C, Surawicz CM (2009) Emerging infections of the gastrointestinal tract. Best Practice & Research Clin Gastroenterol 23:89–99
Schuffenecker I, Ando T, Thouvenot D, Lina B, Aymard M (2001) Genetic classification of “Sapporo-like viruses”. Arch Virol 146:2115–2132
Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34:126–129
Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5:233–241
Svraka S, Vennema H, van der Veer B, Hedlund KO, Thorhagen M, Siebenga J, Duizer E, Koopmans M (2010) Epidemiology and genotype analysis of emerging sapovirus associated infections across Europe. J Clin Microbiol 48:2191–2198
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr121
Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H (2003) Detection of norovirus (GI, GII), Sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J Virol Methods 114:37–44
Xavier MPTP, Oliveira SA, Ferreira MSR, Victoria M, Miranda V, Silva MFM, Strina A, Barreto ML, Miagostovicht MP, Leite JPG (2009) Detection of caliciviruses associated with acute infantile gastroenteritis in Salvador, an urban center in Northeast Brazil. Braz J Med Bio Res 42:438–444
Acknowledgments
The first author is supported financially by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). The fourth and fifth authors are research fellows of CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). We thank Dr. Tomoichiro Oka (National Institute of Infectious Diseases, Japan) for the technical discussion.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2011_1079_MOESM1_ESM.ppt
Supplementary Fig. 1 Schematic representation of the sapovirus genome and the position of primers used for RT-PCR, showing overlapping regions of the clones. Supplementary material 1 (PPT 115 kb)
705_2011_1079_MOESM2_ESM.ppt
Supplementary Fig. 2 Recombination analysis based on the 3′ region of the sapovirus genome using the RDP method. A schematic representation of the genomic region used in this analysis is shown below, to scale, with the corresponding nucleotide positions. Longer boxes represent the genome, and the predicted recombinant regions are shown as smaller boxes below each genome. The possible parental or daughter sequences are indicated by dotted lines. The genogroup-genotype classification is indicated at the right. p-values identified by RDP for each recombination event are indicated below the virus identifier. Supplementary material 2 (PPT 162 kb)
Rights and permissions
About this article
Cite this article
dos Anjos, K., Lima, L.M.P., Silva, P.A. et al. The possible molecular evolution of sapoviruses by inter- and intra-genogroup recombination. Arch Virol 156, 1953–1959 (2011). https://doi.org/10.1007/s00705-011-1079-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1079-8