Abstract
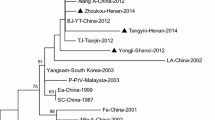
The Chinese hog cholera lapinized virus (HCLV), also called the “Chinese strain” or “C-strain” of classical swine fever virus (CSFV), was developed in China in the 1950s. There are uncertainties about the genetic heterogeneity and origin of this vaccine virus. The objectives of this study were to investigate the genetic heterogeneity of the C-strain, for which nucleotide sequences have been submitted to GenBank from different laboratories, and to determine whether there is any evidence to support the hypothesis that the C-strain originated from the Shimen strain. Analysis of 150 nearly complete E2 gene sequences revealed that the C-strain clade includes several HCLV vaccine strains, cell-culture-adapted Riems strains, and viruses isolated from diseased pigs. The whole-genome phylogeny indicated that the ancestor of the C-strain was only distantly related to the Shimen strain. Therefore, there was no phylogenetic evidence to support the Shimen-origin hypothesis.

Similar content being viewed by others
References
Aynaud JM, Lejolly JC, Bibard C, Galicher C (1971) Studies of the properties of cold induced classical swine fever virus mutants. Application to vaccination. Bull Off Int Epizoot 75:654–659 (in French)
Björklund H, Lowings P, Stadejek T, Vilcek S, Greiser-Wilke I, Paton D, Belák S (1999) Phylogenetic comparison and molecular epidemiology of classical swine fever virus. Virus Genes 19:189–195
Björklund HV, Stadejek T, Vilcek S, Belák S (1998) Molecular characterization of the 3′ noncoding region of classical swine fever virus vaccine strains. Virus Genes 16:307–312
Bognár K, Mészáros J (1963) Experiences with a “Chinese” strain of lapinized swine fever virus. Acta Vet Hung 13:69–74
Chen N, Hu H, Zhang Z, Shuai J, Jiang L, Fang W (2008) Genetic diversity of the envelope glycoprotein E2 of classical swine fever virus: recent isolates branched away from historical and vaccine strains. Vet Microbiol 127:286–299
Dong X, Chen Y (2007) Marker vaccine strategies and candidate CSFV marker vaccines. Vaccine 25:205–230
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Lee RCT, Lai SS (1980) Pathological pictures of rabbits and pigs infected with two Lapinized Hog Cholera viruses. J Chin Soc Vet Sci 6:113–121 (in Chinese)
Leifer I, Depner K, Blome S, Le Potier MF, Le Dimna M, Beer M, Hoffmann B (2009) Differentiation of C-strain “Riems” or CP7_E2alf vaccinated animals from animals infected by classical swine fever virus field strains using real-time RT-PCR. J Virol Methods 158:114–122
Leifer I, Everett H, Hoffmann B, Sosan O, Crooke H, Beer M, Blome S (2010) Escape of classical swine fever C-strain vaccine virus from detection by C-strain specific real-time RT-PCR caused by a point mutation in the primer-binding site. J Virol Methods 166:98–100
Lin TTC, Lee RCT (1981) An overall report on the development of a highly safe and potent lapinized hog cholera virus strain for hog cholera control in Taiwan. Natl Sci Coun Spec Publ 5:1–44
Liu L, Xia H, Baule C, Belák S, Wahlberg N (2010) Effects of methodology and analysis strategy on robustness of pestivirus phylogeny. Virus Res 147:47–52
Liu L, Xia H, Belák S, Widén F (2009) Development of a novel real-time PCR assay based on primer-probe energy transfer for detection of classical swine fever virus. J Virol Methods 160:69–73
Liu L, Xia H, Wahlberg N, Belák S, Baule C (2009) Phylogeny, classification and evolutionary insights into pestiviruses. Virology 385:351–357
Oleksiewicz MB, Rasmussen TB, Normann P, Uttenthal A (2003) Determination of the sequence of the complete open reading frame and the 5′NTR of the Paderborn isolate of classical swine fever virus. Vet Microbiol 92:311–325
Paton DJ, McGoldrick A, Greiser-Wilke I, Parchariyanon S, Song JY, Liou PP, Stadejek T, Lowings JP, Björklund H, Belák S (2000) Genetic typing of classical swine fever virus. Vet Microbiol 73:137–157
Research Group of CSF Vaccine (1979) Studies on the avirulent Lapinized Hog Cholera virus. Acta Vet Zootech Sin 10:1–34 (in Chinese)
Sasahara J, Kumagai T, Shimizu Y, Furuuchi S (1969) Field experiments of hog cholera live vaccine prepared in guinea-pig kidney cell culture. Natl Inst Anim Health Q (Tokyo) 9:83–91
Suradhat S, Kesdangsakonwut S, Sada W, Buranapraditkun S, Wongsawang S, Thanawongnuwech R (2006) Negative impact of porcine reproductive and respiratory syndrome virus infection on the efficacy of classical swine fever vaccine. Vaccine 24:2634–2642
Terpstra C, de Smit AJ (2000) The 1997/1998 epizootic of swine fever in the Netherlands: control strategies under a non-vaccination regimen. Vet Microbiol 77:3–15
Thiel H-J, Collett MS, Gould EA, Heinz FX, Houghton M, Meyers G, Purcell RH, Rice CM (2005) Family Flaviviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy. VIIIth report of the ICTV. Elsevier/Academic Press, London, pp 979–996
Vilcek S, Stadejek T, Ballagi-Pordány A, Lowings JP, Paton DJ, Belák S (1996) Genetic variability of classical swine fever virus. Virus Res 43:137–147
Wu HX, Wang JF, Zhang CY, Fu LZ, Pan ZS, Wang N, Zhang PW, Zhao WG (2001) Attenuated lapinized Chinese strain of classical swine fever virus: complete nucleotide sequence and character of 3’-noncoding region. Virus Genes 23:69–76
Xiao M, Gao J, Wang Y, Wang X, Lu W, Zhen Y, Chen J, Li B (2004) Influence of a 12-nt insertion present in the 3’ untranslated region of classical swine fever virus HCLV strain genome on RNA synthesis. Virus Res 102:191–198
Zhang XJ, Xia H, Everett H, Sosan O, Crooke H, Belák S, Qiu HJ, Liu L (2010) Evaluation of a primer-probe energy transfer real-time PCR assay for detection of classical swine fever virus. J Virol Methods 168:259–261
Zhao JJ, Cheng D, Li N, Sun Y, Shi Z, Zhu QH, Tu C, Tong GZ, Qiu HJ (2008) Evaluation of a multiplex real-time RT-PCR for quantitative and differential detection of wild-type viruses and C-strain vaccine of classical swine fever virus. Vet Microbiol 126:1–10
Zhou T (1980) Classical swine fever virus and progresses in its control (part I). Chin J Vet Sci Technol 4:23–33 (in Chinese)
Zhou T (1980) Classical swine fever virus and progresses in its control (part II). Chin J Vet Sci Technol 5:30–39 (in Chinese)
Acknowledgements
We are grateful to Prof. Mészáros, Veterinary Research Institute of the Hungarian Academy of Sciences, Budapest, Hungary, for his fruitful comments on the manuscript. The work was supported by the Award of Excellence (Excellensbidrag) provided to SB by the Swedish University of Agricultural Sciences (SLU), Sweden.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xia, H., Wahlberg, N., Qiu, HJ. et al. Lack of phylogenetic evidence that the Shimen strain is the parental strain of the lapinized Chinese strain (C-strain) vaccine against classical swine fever. Arch Virol 156, 1041–1044 (2011). https://doi.org/10.1007/s00705-011-0948-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-0948-5