Abstract
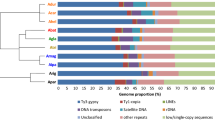
Satellite DNA is a class of tandemly repetitive sequences characterized by rapid evolution and high intra- and inter-specific diversification and is usually the major component of the constitutive heterochromatin. We investigated the abundance of the satDNA in 77 species of Asclepias from Americas, a genus of the Apocynaceae family. De novo identification of the most abundant satDNA sequences was performed using Tandem Repeat Analyzer and Repeat Explorer pipelines. The abundance of satDNA in 77 species was determined using mapping reads, and the correlation between the abundance of satDNAs and clades on the phylogeny tree was estimated by lambda statistics using phytools R package. The repetitive sequences in Asclepias were mostly constitutive by transposable elements, ranged 11.11 to 16.32%. The satDNA ranged 0.985 to 7.732% and we obtained 12 satDNA sequences (call Sat1 to Sat12), being for the Sat1 we obtained seven families distributed across all species. The Sat2, Sat3, and families of Sat1 showed significant correlations with the topologies of the phylogenetic tree, among which some families of satDNAs displayed varying abundances across the main clades. We concluded that Asclepias is characterized by many satDNAs, and that the abundance of satDNAs is in accordance with the phylogeny and geographical distribution.

Similar content being viewed by others
Data availability
All data analyzed during this study are included in Online Resource 3 supplementary information.
References
Almeida C, Fonsêca A, Santos KGB, Mosiolek M, Pedrosa-Harand A (2012) Contrasting evolution of a satellite DNA and its ancestral IGS rDNA in Phaseolus (Fabaceae). Genome 55:683–689. https://doi.org/10.1139/g2012-059
Anamthawat-Jónsson K, Heslop-Harrison JS (1992) Species specific DNA sequences in the Triticeae. Hereditas 116:49–54. https://doi.org/10.1111/j.1601-5223.1992.tb00204.x
Bai C, Alverson WS, Follansbee A, Waller DM (2012) New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot (Oxford) 110:1623–1629. https://doi.org/10.1093/aob/mcs222
Belyayev A, Josefiová J, Jandová M, Kalendar R, Krak K, Mandák B (2019) Natural history of a satellite DNA family: from the ancestral genome component to species-specific sequences, concerted and non-concerted evolution. Int J Molec Sci 20:1201. https://doi.org/10.3390/ijms20051201
Broyles SB, Elkins G (2019) Hybridization between Asclepias purpurascens and Asclepias syriaca (Apocynaceae): a cause for concern? J Torrey Bot Soc 146:278–290. https://doi.org/10.3159/TORREY-D-18-00050.1
Chuba D, Goyder D, Chase MW, Fishbein M (2017) Phylogenetics of the African Asclepias complex (Apocynaceae) based on three plastid DNA regions. Syst Bot 42:148–159. https://doi.org/10.1600/036364417X694539
Fishbein M, Chuba D, Ellison C, Mason-Gamer RJ, Lynch SP (2011) Phylogenetic relationships of Asclepias (Apocynaceae) inferred from non-coding chloroplast DNA sequences. Syst Bot 36:1008–1023. https://doi.org/10.1600/036364411x605010
Fishbein M, Straub SCK, Boutte J, Hansen K, Cronn RC, Liston A (2018) Evolution at the tips: Asclepias phylogenomics and new perspectives on leaf surfaces. Amer J Bot 105:514–524. https://doi.org/10.1002/ajb2.1062
Gill N, Buti M, Kane N, Bellec A, Helmstetter N, Berges H, Rieseberg LH (2014) Sequence-based analysis of structural organization and composition of the cultivated sunflower (Helianthus annuus L.) Genome. Biology (Basel) 3:295–319. https://doi.org/10.3390/biology3020295
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molec Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nature Meth 9:357–357. https://doi.org/10.1038/nmeth.1923
Macas J, Mészáros T, Nouzová M (2002) PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18:28–35. https://doi.org/10.1093/bioinformatics/18.1.28
Macas J, Novak P, Pellicer J, Čížková J, Koblížková A, Neumann P, Fuková I, Doležel J, Kelly LJ, Leitch IJ (2015) In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe fabeae. PLoS ONE 10:1–23. https://doi.org/10.1371/journal.pone.0143424
Marçais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. https://doi.org/10.1093/bioinformatics/btr011
Mattila HR, Otis GW (2003) A comparison of the host preference of monarch butterflies (Danaus plexippus) for milkweed (Asclepias syriaca) over dog-strangler vine (Vincetoxicum rossicum). Entomol Exp Appl 107:193–199. https://doi.org/10.1046/j.1570-7458.2003.00049.x
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genomics Proteomics Bioinf 12:164–171. https://doi.org/10.1016/j.gpb.2014.07.003
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mole Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Novak P, Neumann P, Macas J (2010) Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinf 11:378
Novak P, Robledillo LA, Koblizkova A, Vrbova I, Neumann P, Macas J (2017) TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucl Acids Res 45:e111
Oliveira MAS, Nunes T, Santos MA, Gomes DF, Costa I, Van-Lume B, Silva SSM, Oliveira RS, Simon MF, Lima GSA, Gissi DS, Almeida CCS, Souza G, Marques A (2021) High-throughput genomic data reveal complex phylogenetic relationships in Stylosanthes Sw (Leguminosae). Frontiers Genet 12:727314. https://doi.org/10.3389/fgene.2021.727314
Rabinowicz PD, Bennetzen JL (2006) The maize genome as a model for efficient sequence analysis of large plant genomes. Curr Opin Pl Biol 9:149–156. https://doi.org/10.1016/j.pbi.2006.01.015
Rebollo R, Romanish MT, Mager DL (2012) Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annul Rev Genet 46:21–42. https://doi.org/10.1146/annurev-genet-110711-155621
Revell LJ (2012) phytools: an R package for phylogenetic comparative biology (and other things). Meth Ecol Evol 3:217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x
Sarmashghi S, Balaban M, Rachtman E, Touti B, Mirarab S, Bafna V (2021) Estimating repeat spectra and genome length from low-coverage genome skims with RESPECT. PLoS Comput Biol 17:e1009449. https://doi.org/10.1371/journal.pcbi.1009449
Souza MCP, Silva JN, Almeida C (2013) Differential detection of transposable elements between Saccharum species. Gen Molec Biol 36:408–412. https://doi.org/10.1590/S1415-47572013005000030
Straub SC, Fishbein M, Livshultz T, Foster Z, Parks M, Weitemier K, Cronn RC, Liston A (2011) Building a model: developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genom 12:211. https://doi.org/10.1186/1471-2164-12-211
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. https://doi.org/10.1038/35048692
Vitales D, Garcia S, Dodsworth S (2020) Reconstructing phylogenetic relationships based on repeat sequence similarities. Molec Phylogen Evol 147:106766. https://doi.org/10.1016/j.ympev.2020.106766
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982. https://doi.org/10.1038/nrg2165
Acknowledgements
We would like to thank the Federal University of Alagoas for the access to laboratories and scientific support, the Fundação de Amparo à Pesquisa de Alagoas (FAPEAL) for funding this project, and the National Council for the Improvement of Higher Education (CAPES).
Author information
Authors and Affiliations
Contributions
G.C.C. conceived the project and the experiments. C.A. drafted the manuscript. All authors contributed to the final manuscript draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Ethical approval
Not applicable.
Additional information
Handling Editor: Ales Kovarik.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Throughput sequencing data of the Asclepias used in the study.
Online Resource 2. Maximum likelihood phylogeny obtained using concatenated chloroplast regions and support estimated by bootstraps (in percentages).
Online Resource 3. Monomer sequences of satellite DNA in Asclepias.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Costa, G.C., Almeida, C. Identification of differential abundance of satellite DNA sequences in Asclepias (Apocynaceae): in-depth characterization of species-specific sequences. Plant Syst Evol 308, 41 (2022). https://doi.org/10.1007/s00606-022-01832-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-022-01832-0