Abstract
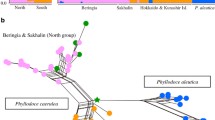
Geranium erianthum is an alpine plant growing in dry habitats, which is distributed from East Asia to northern coastal regions of the northern Pacific. The ice-free area around the current Bering Strait (i.e., Beringia) had played an important role in range expansion into neighboring regions such as East Asia and North America for some alpine plants. However, recent studies suggest that some alpine plants in snowbed environment spread from East Asia to northern coastal regions of the northern Pacific. In this study, we investigated phylogenetic relationships and genetic differentiations among populations of G. erianthum and the related species using the chloroplast genome and single-nucleotide polymorphisms, to evaluate the alternative biogeographic hypotheses in which region of Beringia, British Columbia or East Asia is probable for its distributional origin. Range reconstruction based on phylogenetic tree of chloroplast genome indicated G. erianthum and related species originated in East Asia, from where G. erianthum migrated eastward into Beringia and British Columbia. In addition, nuclear genome-wide SNPs indicated that no significant genetic differentiation was detected between Japanese and Beringian populations. The lack of genetic differentiation suggests that the current range of G. erianthum resulted from rapid range expansion during the latter period of the last glacial era. Overall, the East Asian refugium hypothesis was applicable to the alpine plant G. erianthum in dry habitat, indicating that range expansion pattern from East Asia into the northern Pacific may be more common rather than limited for snowbed species.




Similar content being viewed by others
Availability of data and material
The chloroplast genome sequence results are deposited in DDBJ (Accession numbers: DRA010649). SNPs data from MIG-seq analysis are also deposited in DDBJ (Accession numbers: DRA010651).
References
Aedo C (2001) The genus Geranium L. (Geraniaceae) in North America II. Perennial Species. Anales Jard Bot Madrid 58:39–82. https://doi.org/10.3989/ajbm.2001.v59.i1.74
Aedo C (2017) Revision of Geranium (Geraniaceae) in the western and central pacific area. Syst Bot Monogr 102:1–240
Akiyama S (2001) Geraniaceae. In: Iwatsuki K, Boufford DE, Ohba H (eds) Flora of Japan Ib. Kodansha Ltd., Tokyo, pp 287–293
Alsos IG, Engelskjon T, Gielly L, Taberlet P, Brochmann C (2005) Impact of ice ages on circumpolar molecular diversity: insights from an ecological key species. Molec Ecol 14:2739–2753. https://doi.org/10.1111/j.1365-294X.2005.02621.x
Arenas M, Ray N, Currat M, Excoffier L (2012) Consequences of range contractions and range shifts on molecular diversity. Molec Biol Evol 29:207–218. https://doi.org/10.1093/molbev/msr187
Asikainen E, Mutikainen P (2005) Preferences of pollinators and herbivores in gynodioecious Geranium sylvaticum. Ann Bot (Oxford) 95:879–886. https://doi.org/10.1093/aob/mci094
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376. https://doi.org/10.1371/journal.pone.0003376
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. J Bioinf 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Molec Ecol 22:3124–3140. https://doi.org/10.1111/mec.12354
Chang SM (2007) Gender-specific inbreeding depression in a gynodioecious plant, Geranium maculatum (Geraniaceae). Amer J Bot 94:1193–1204. https://doi.org/10.3732/ajb.94.7.1193
Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T (2020) ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Molec Biol Evol 37:291–294. https://doi.org/10.1093/molbev/msz189
Davis MB, Shaw RG (2001) Range shifts and adaptive responses to quaternary climate change. Science 292:673–679. https://doi.org/10.1126/science.292.5517.673
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res 45:e18. https://doi.org/10.1093/nar/gkw955
Dlusskiĭ GM, Lavrova NV, Erofeeva EA (2000) The mechanisms of the restriction of pollinator range in the fireweed (Chamaenerion angustifolium) and 2 species of Geranium (Geranium palustre) and G. pratense). Zhurn Obshchei Biol 61:181–197
Dobson M (1994) Patterns of distribution in Japanese land mammals. Mammal Rev 24:91–111. https://doi.org/10.1111/j.1365-2907.1994.tb00137.x
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. https://doi.org/10.1186/1471-2148-7-214
Ehrich D, Gaudeul M, Assefa A, Koch MA, Mummenhoff K, Nemomissa S, Consortium I, Brochmann C (2007) Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molec Ecol 16:2542–2559. https://doi.org/10.1111/j.1365-294X.2007.03299.x
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Molec Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164:1567–1587. https://doi.org/10.1093/genetics/164.4.1567
Fiz O, Vargas P, Alarcon M, Aedo C, Garcia JL, Aldasoro JJ (2008) Phylogeny and historical biogeography of Geraniaceae in relation to climate changes and pollination ecology. Syst Bot 33:326–342. https://doi.org/10.1600/036364408784571482
Frenzel B (1968) The Pleistocene vegetation of northern Eurasia. Science 161:637–649. https://doi.org/10.1126/science.161.3842.637
Fujii N, Ueda K, Watano Y, Shimizu T (1997) Intraspecific sequence variation of chloroplast DNA in Pedicularis chamissonis Steven (Scrophulariaceae) and geographic structuring of the Japanese “Alpine” plants. J Pl Res 110:195–207. https://doi.org/10.1007/bf02509308
Fujii N, Ueda K, Watano Y, Shimizu T (1999) Further analysis of intraspecific sequence variation of chloroplast DNA in Primula cuneifolia Ledeb. (Primulaceae): Implications for biogeography of the Japanese alpine flora. J Pl Res 112:87–95. https://doi.org/10.1007/pl00013866
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Molec Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Hampe A, Pemonge MH, Petit RJ (2013) Efficient mitigation of founder effects during the establishment of a leading-edge oak population. Proc Roy Soc B Biol Sci 280:20131070. https://doi.org/10.1098/rspb.2013.1070
Hata D, Higashi H, Yakubov V, Barkalov V, Ikeda H, Setoguchi H (2017) Phylogeographical insight into the Aleutian flora inferred from the historical range shifts of the alpine shrub Therorhodion camtschaticum (Pall.) Small (Ericaceae). J Biogeogr 44:283–293. https://doi.org/10.1111/jbi.12876
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276. https://doi.org/10.1111/j.1095-8312.1996.tb01434.x
Hewitt GM (2000) The genetic legacy of the quaternary ice ages. Nature 405:907–913. https://doi.org/10.1038/35016000
Hirano T, Saito T, Tsunamoto Y, Koseki J, Ye B, Do VT, Miura O et al (2019) Enigmatic incongruence between mtDNA and nDNA revealed by multi-locus phylogenomic analyses in freshwater snails. Sci Rep 9:6223. https://doi.org/10.1038/s41598-019-42682-0
Hopkins DM (1967) The Bering land bridge. Stanford University Press, Stanford
Hultén E (1937) Outline of the history of Arctic and Boreal Biota during the quaternary period. Bokförlags aktiebolaget Thule, Stockholm
Ikeda H, Yakubov V, Barkalov V, Setoguchi H (2018) Post-glacial East Asian origin of the alpine shrub Phyllodoce aleutica (Ericaceae) in Beringia. J Biogeogr 45:1261–1274. https://doi.org/10.1111/jbi.13230
Ikeda H, Yakubov V, Barkalov V, Sato K, Fujii N (2020) East Asian origin of the widespread alpine snow-bed herb, Primula cuneifolia (Primulaceae), in the northern Pacific region. J Biogeogr 47:2181–2193. https://doi.org/10.1111/jbi.13918
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kadota Y (2016) Geraniaceae. In: Ohashi H, Kadota Y, Murata J, Yonekura K, Kihara H (eds) Wild Flowers of Japan, vol 3. Heibonsha, Tokyo, pp 248–253 (in Japanese)
Kandori I (2002) Diverse visitors with various pollinator importance and temporal change in the important pollinators of Geranium thunbergii (Geraniaceae). Ecol Res 17:283–294. https://doi.org/10.1046/j.1440-1703.2002.00488.x
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molec Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Koidzumi G (1919) Genetic and floristic phytogeography of the alpine flora of Japan. Shokubutsugaku Zasshi 33:193–222. https://doi.org/10.15281/jplantres1887.33.393_193
Korotky A, Grebennikova T, Razjigaeva N, Volkov V, Mokhova L, Ganzey L, Bazarova V (1997) Marine terraces of western Sakhalin island. Catena 30:61–81. https://doi.org/10.1016/s0341-8162(97)00002-7
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. J Bioinf 35:4453–4455. https://doi.org/10.1093/bioinformatics/btz305
Kurata S, Sakaguchi S, Hirota SK, Kurashima O, Suyama Y, Nishida S, Ito M (2021) Refugia within refugium of Geranium yesoense (Geraniaceae) in Japan were driven by recolonisation into the southern interglacial refugium. Biol J Linn Soc 132:552–572. https://doi.org/10.1093/biolinnean/blaa212
Lassmann T, Hayashizaki Y, Daub CO (2009) TagDust-a program to eliminate artifacts from next generation sequencing data. J Bioinf 25:2839–2840. https://doi.org/10.1093/bioinformatics/btp527
Lelej AS, Kupianskaya AN (2000) The Bumble-bees (Hymenoptera, Apidae, Bombinae) of the Kuril Islands. Far E Entomol 95:1–17
Matenaar D, Fingerle M, Heym E, Wirtz S, Hochkirch A (2018) Phylogeography of the endemic grasshopper genus Betiscoides (Lentulidae) in the South African cape floristic region. Molec Phylogen Evol 118:318–329. https://doi.org/10.1016/j.ympev.2017.09.024
Matzke N (2013) Probabilistic historical biogeography: New models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Frontiers Biogeogr 5:242–248. https://doi.org/10.21425/F5FBG19694
McKay BD (2012) A new timeframe for the diversification of Japan’s mammals. J Biogeogr 39:1134–1143. https://doi.org/10.1111/j.1365-2699.2011.02666.x
Michalski SG, Durka W (2012) Assessment of provenance delineation by genetic differentiation patterns and estimates of gene flow in the common grassland plant Geranium pratense. Conservation Genet 13:581–592. https://doi.org/10.1007/s10592-011-0309-7
Milligan B (1992) Plant DNA isolation. In: Hoelzel AR (ed) Molecular genetic analysis of populations: a practical approach. IRL Press, Oxford, pp 59–88
Nazareno AG, Bemmels JB, Dick CW, Lohmann LG (2017) Minimum sample sizes for population genomics: an empirical study from an Amazonian plant species. Molec Ecol Resources 17:1136–1147. https://doi.org/10.1111/1755-0998.12654
Newton AC, Allnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145. https://doi.org/10.1016/s0169-5347(98)01555-9
Okuyama Y, Goto N, Nagano AJ, Yasugi M, Kokubugata G, Kudoh H, Qi ZC, Ito T, Kakishima S, Sugawara T (2020) Radiation history of Asian Asarum (sect. Heterotropa, Aristolochiaceae) resolved using a phylogenomic approach based on double-digested RAD-seq data. Ann Bot (Oxford) 126:245–260. https://doi.org/10.1093/aob/mcaa072
Ota Y, Machida H (1987) Quaternary sea-level changes in Japan. In: Tooley MJ, Shennan I (eds) Sea-level changes. The Institute of British geographers special publications series 20. Blackwell, Basil, pp 182–224
Palazzesi L, Gottschling M, Barreda V, Weigend M (2012) First Miocene fossils of Vivianiaceae shed new light on phylogeny, divergence times, and historical biogeography of Geraniales. Biol J Linn Soc 107:67–85. https://doi.org/10.1111/j.1095-8312.2012.01910.x
Park S, Ruhlman TA, Weng ML, Hajrah NH, Sabir JSM, Jansen RK (2017) Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of Geranium. Genome Biol Evol 9:1766–1780. https://doi.org/10.1093/gbe/evx124
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molec Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Amer J Human Genet 81:559–575. https://doi.org/10.1086/519795
R Core Team (2019) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Available at: https://www.R-project.org/
Razjigaeva NG, Ganzey LA, Belyanina NI, Grebennikova TA, Ganzey KS (2008) Paleoenvironments and landscape history of the minor Kuril Islands since the late glacial. Quatern Int 179:83–89. https://doi.org/10.1016/j.quaint.2007.10.017
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Schönswetter P, Tribsch A, Stehlik I, Niklfeld H (2004) Glacial history of high alpine Ranunculus glacialis (Ranunculaceae) in the European Alps in a comparative phylogeographical context. Biol J Linn Soc 81:183–195. https://doi.org/10.1111/j.1095-8312.2003.00289.x
Shi MM, Chen XY (2012) Leading-edge populations do not show low genetic diversity or high differentiation in a wind-pollinated tree. Populat Ecol 54:591–600. https://doi.org/10.1007/s10144-012-0332-7
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. J Bioinf 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stewart JR, Lister AM, Barnes I, Dalen L (2010) Refugia revisited: individualistic responses of species in space and time. Proc Roy Soc London Ser B Biol Sci 277:661–671. https://doi.org/10.1098/rspb.2009.1272
Suyama Y, Matsuki Y (2015) MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci Rep 5:16963. https://doi.org/10.1038/srep16963
Takahashi Y, Suyama Y, Matsuki Y, Funayama R, Nakayama K, Kawata M (2016) Lack of genetic variation prevents adaptation at the geographic range margin in a damselfly. Molec Ecol 25:4450–4460. https://doi.org/10.1111/mec.13782
Tanabe AS (2008) Phylogears. Ver. 2.0. Available at: https://www.fifthdimension.jp/products/phylogears/. Accessed 20 Nov 2021
Terossi M, De Grave S, Mantelatto FL (2017) Global biogeography, cryptic species and systematic issues in the shrimp genus Hippolyte Leach, 1814 (Decapoda: Caridea: Hippolytidae) by multimarker analyses. Sci Rep 7:6697. https://doi.org/10.1038/s41598-017-06756-1
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq - versatile and accurate annotation of organelle genomes. Nucl Acids Res 45:W6–W11. https://doi.org/10.1093/nar/gkx391
Tsukada M (1988) Glacial and holocene vegetation history of Japan. In: Huntley B, Webb T (eds) Handbook of Vegetation Science 7. Vegetation history. Kluwer, Dordrecht, pp 459–518
Wakasugi Y, Azuma H, Naiki A, Nishida S (2017) Morphological and molecular phylogenetic analyses of Geranium yesoense (Geraniaceae) in Japan. Acta Phytotax Geobot 68:129–144. https://doi.org/10.18942/apg.201708
Watanabe Y, Kawamata I, Matsuki Y, Suyama Y, Uehara K, Ito M (2018) Phylogeographic analysis suggests two origins for the riparian azalea Rhododendron indicum (L.) Sweet. Heredity 121:594–604. https://doi.org/10.1038/s41437-018-0064-3
Williams CF, Kuchenreuther MA, Drew A (2000) Floral dimorphism, pollination, and self-fertilization in gynodioecious Geranium richardsonii (Geraniaceae). Amer J Bot 87:661–669. https://doi.org/10.2307/2656852
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058. https://doi.org/10.1073/pnas.84.24.9054
Xu L, Aedo C (2008) Flora of China, vol 11. Science Press Missouri Botanical Garden Press, St. Louis
Ye JW, Guo XD, Wang SH, Bai WN, Bao L, Wang HF, Ge JP (2015) Molecular evidence reveals a closer relationship between Japanese and mainland subtropical specimens of a widespread tree species, Acer mono. Biochem Syst Ecol 60:14–149. https://doi.org/10.1016/j.bse.2015.04.010
Yonekura K (2005) Taxonomic Notes on vascular plants in japan and its adjacent regions (I). New combinations and new names of Japanese plants. J Jap Bot 80:323–333
Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, Balfour J (2000) Molecular analysis of plant migration and refugia in the Arctic. Science 289:1343–1346. https://doi.org/10.1126/science.289.5483.1343b
Acknowledgements
The authors thank Dr. Kenji Horie, Dr. Naoyuki Nakahama, Dr. Sachiko Nishida, and Tomohiro Kawai for granting access to their collection of materials. We also thank the Ministry of the Environment, and the prefectural offices of Hokkaido, Nagano, and Yamanashi for granting us permission to conduct surveys. We thank The University Museum, University of Tokyo (TI) for granting access to their specimen collections.
Funding
This study was partly supported by JSPS KAKENHI Grant Number JP16H04827 and the National BioResource Project (18km0210136j0002) from AMED.
Author information
Authors and Affiliations
Contributions
SK, SS, HI and MI to conceived the ideas; SKH and YS performed MIG-seq experiment; OK supported to detect genotype and SNP calling from next-generation sequencing data; SK and SS designed the study and genetic analyses. All authors contributed to interpret the analysis results and write the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
All authors consent to participate in this manuscript.
Consent for publication
All authors consent for publication of this manuscript.
Additional information
Handling Editor: Christian Parisod.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. DDBJ and voucher specimen numbers for chloroplast genome sequencing.
Online Resource 2. Voucher specimen numbers for MIG-seq analyses.
Online Resource 3. Nucleotide substitution models in RAxML-NG, MrBayes, and BEAST.
Online Resource 4. Sequence alignments of 40 taxa chloroplast genome. The 68 coding sequences are concatenated.
Online Resource 5. Bayesian inference tree based on the chloroplast genome. Numbers above branches are posterior probabilities.
Online Resource 6. Results of SNPs detection of Geranium erianthum, which are based on MIG-seq analysis of the nuclear genome.
Online Resource 7. ΔK and mean estimated LnP(D) in STRUCTURE analysis.
Online Resource 8. Accession numbers of Geraniaceae and Francoaceae species.
Rights and permissions
About this article
Cite this article
Kurata, S., Sakaguchi, S., Ikeda, H. et al. From East Asia to Beringia: reconstructed range dynamics of Geranium erianthum (Geraniaceae) during the last glacial period in the northern Pacific region. Plant Syst Evol 308, 28 (2022). https://doi.org/10.1007/s00606-022-01820-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-022-01820-4