Abstract
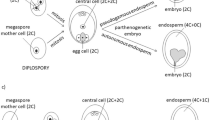
Apomixis is the ability of plants to produce asexual seeds, which are clones of the mother plant. The phenomenon of apomixis is tightly linked to ploidy, where diploids lack apomixis and reproduce sexually, while higher ploidy levels can exhibit apomixis. Taraxacum F.H.Wigg. species (dandelions) commonly exhibit apomixis; however, only limited studies have evaluated genome size variation of the iconic weedy dandelion, Taraxacum officinale F.H.Wigg. (common dandelion), particularly in North America. To evaluate the ploidy and reproductive biology of common dandelion, we created a global collection, with an emphasis on North America and determined genome size by evaluating seeds with flow cytometry. A total of 635 accessions were screened, and with the exception of two accessions from Germany and Austria, all were found to exclusively contain polyploid seed. The mode of reproduction of a sample of 96 of these accessions was observed by conducting emasculations. Our results indicate that the diploid, sexual cytotype of common dandelion may be absent in North America, while diploids were found in previously described areas of Central Europe. This suggests that the clonal lineages of common dandelion in North America may largely be derivative from apomictic plants introduced from abroad. Furthermore, if all North American common dandelion is apomictic, it may be unreceptive to pollen, which may isolate it from other dandelion species and inform the potential for gene flow.



Similar content being viewed by others
References
Andersson TN, Milberg P (1998) Weed flora and the relative importance of site, crop, crop rotation, and nitrogen. Weed Sci 46:30–38
Bennett MD, Smith JB, Smith RIL (1982) DNA amounts of angiosperms from the Antarctic and South Georgia. Environm Exp Bot 22:307–318. doi:10.1016/0098-8472(82)90023-5
Bowers JE (1990) Natural rubber-producing plants for the United States. National Agricultural Library, Beltsville
Brock MT (2004) The potential for genetic assimilation of a native dandelion species, Taraxacum ceratophorum (Asteraceae), by the exotic congener T. officinale. Amer J Bot 91:656–663. doi:10.3732/ajb.91.5.656
den Nijs JCM, Kirschner J, Štěpánek J, van der Hulst A (1990) Distribution of diploid sexual plants of Taraxacum sect. Ruderalia in east-central Europe, with special reference to Czechoslovakia. Pl Syst Evol 170:71–84. doi:10.1007/BF00937850
Doležel J, Bartoš J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot (Oxford) 95:99–110. doi:10.1093/aob/mci005
Doll R (1982) Grundriß der evolution der gattung Taraxacum Zinn. Feddes Repert 93:481–624. doi:10.1002/fedr.19820930702
Free JB (1968) Dandelion as a competitor to fruit trees for bee visits. J Appl Ecol 5:169–178. doi:10.2307/2401281
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220:1049–1051. doi:10.1126/science.220.4601.1049
Galbraith DW, Lambert GM, Macas J, Dolezel J (2001) Analysis of nuclear DNA content and ploidy in higher plants. In: Robinson JP, Darzynkiewicz Z, Dean PN, Dressler LG, Orfao A, Rabinovitch PS, Stewart CC, Tanke HJ, Wheeless LL (eds) Current protocols in cytometry. Wiley, New York, pp 7.6.1–7.6.22. doi:10.1002/0471142956.cy0706s02
Gyenes V, Béres I (2006) The allelopathic potential of common dandelion (Taraxacum officinale WEB.). J Pl Diseases Protect New Ser 20:471–478
Integrated Taxonomic Information System online database (2017). Available at: http://www.itis.gov. Accessed 28 Jan 2017
Kapusta G, Krausz RF (1993) Weed control and yield are equal in conventional, reduced-, and no-tillage soybean (Glycine max) after 11 years. Weed Technol 7:443–451. http://www.jstor.org/stable/3987627
Kirschner J, Štěpánek J (1996) Modes of speciation and evolution of the sections in Taraxacum. Folia Geobot 31:415–426. doi:10.1007/BF02815386
Kirschner J, Štěpánek J (1998) A revision of Taraxacum sect. Piesis (Compositae). Folia Geobot 33:391–414. doi:10.1007/BF02803642
Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, Tonosaki K, Shirasawa S, Fukushima A, Yokoi S, Takahata Y, Kakizaki T, Ishida M, Okamoto S, Sakamoto K, Shirasawa K, Tabata S, Nishio T (2014) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res 21:481–490. doi:10.1093/dnares/dsu014
Luc Brouillet (2006) Taraxacum. In: Flora of North America Editorial Committee (eds) 1993 + , Flora of North America North of Mexico. 20 + vols. New York and Oxford. Vol. 19, 20 and 21, pp 8, 215, 239, 240, 241, 242. Available at: http://www.efloras.org/florataxon.aspx?flora_id=1&taxon_id=132314. Accessed 28 Jan 2017
Lyman JC, Ellstrand NC (1984) Clonal diversity in Taraxacum officinale (compositae), an apomict. Heredity 53:1–10. doi:10.1038/hdy.1984.58
Mártonfiová L (2006) Possible pathways of the gene flow in Taraxacum Sect. Ruderalia. Folia Geobot 41:183–201. doi:10.1007/BF02806478
Menken SBJ, Smit E, den Nijs JCM (1995) Genetical population structure in plants: gene flow between diploid sexual and triploid asexual dandelions (Taraxacum Section Ruderalia). Evolution (Lancaster) 49:1108–1118. doi:10.2307/2410435
Mooibroek H, Cornish K (2000) Alternative sources of natural rubber. Appl Microbiol Biotechnol 53:355–365. doi:10.1007/s002530051627
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. Available at: http://www.R-project.org/. Accessed 18 May 2016
Richards AJ (1970) Hybridization in Taraxacum. New Phytol 69:1103–1121. doi:10.1111/j.1469-8137.1970.tb02492.x
Shibaike H, Akiyama H, Uchiyama S, Kasai K, Morita T (2002) Hybridization between European and Asian dandelions (Taraxacum section Ruderalia and section Mongolica). J Pl Res 115:321–328. doi:10.1007/s10265-002-0045-7
Sliwinska E, Zielinska E, Jedrzejczyk I (2005) Are seeds suitable for flow cytometric estimation of plant genome size? Cytometry A 64A:72–79. doi:10.1002/cyto.a.20122
Sliwinska E, Pisarczyk I, Pawlik A, Galbraith DW (2009) Measuring genome size of desert plants using dry seeds. Botany (Ottawa) 87:127–135. doi:10.1139/B08-120
Solbrig OT (1971) The population biology of dandelions. Amer Sci 59:686–694
Stewart-Wade SM, Neumann S, Collins LL, Boland GJ (2002) The biology of Canadian weeds. 117. Taraxacum officinale G. H. Weber ex Wiggers. Canad J Pl Sci 82:825–853. doi:10.4141/P01-010
Suda J, Krahulcová A, Trávníek P, Krahulec F (2006) Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon 55:447–450. doi:10.2307/25065591
van Baarlen P, van Dijk PJ, Hoekstra RF, de Jong JH (2000) Meiotic recombination in sexual diploid and apomictic triploid dandelions (Taraxacum officinale L.). Genome 43:827–835
van Baarlen P, de Jong HJ, van Dijk PJ (2002) Comparative cyto-embryological investigations of sexual and apomictic dandelions (Taraxacum) and their apomictic hybrids. Sexual Pl Reprod 15:31–38. doi:10.1007/s00497-002-0132-x
van Dijk PJ (2003) Ecological and evolutionary opportunities of apomixis: insights from Taraxacum and Chondrilla. Philos Trans, Ser B 358:1113–1121. doi:10.1098/rstb.2003.1302
van Dijk P, van Damme J (2000) Apomixis technology and the paradox of sex. Trends Pl Sci 5:81–84. doi:10.1016/S1360-1385(99)01545-9
van Dijk P, de Jong HJ, Vijverberg K, Biere A (2009) An apomixis-gene’s view on dandelions. In: Schön I, Martens K, van Dijk P (eds) Lost Sex: The Evolutionary Biology of Parthenogenesis. Springer, Netherlands, pp. 475–493. doi:10.1007/978-90-481-2770-2_22
van Soest JL (1958) Taraxacum Sectio Erythrosperma Dahlstedt Em. Lindberg F. in North-America. Acta Bot Neerl 7:627–628. doi:10.1111/j.1438-8677.1958.tb00635.x
Záveský L, Jarolímová V, Štěpánek J (2005) Nuclear DNA content variation within the genus Taraxacum (asteraceae). Folia Geobot 40:91–104
Zhang Y, Iaffaldano BJ, Xie W, Blakeslee JJ, Cornish K (2015) Rapid and hormone-free Agrobacterium rhizogenes-mediated transformation in rubber producing dandelions Taraxacum kok-saghyz and T. brevicorniculatum. Industr Crops Prod 66:110–118. doi:10.1016/j.indcrop.2014.12.013
Zhang Y, Iaffaldano BJ, Zhuang X, Cardina J, Cornish K (2017) Chloroplast genome resources and molecular markers differentiate rubber dandelion species from weedy relatives. BMC Pl Biol 17:34. doi:10.1186/s12870-016-0967-1
Acknowledgements
We would first like to thank all those who contributed germplasm to this survey, as well as Catherine Herms, who helped assemble the accessions. We are also grateful to Kelsey Redick and Natalie Gustafson for help processing samples. The United States Department of Agriculture, National Institute of Food and Agriculture, Hatch project 230837 partially funded this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Martin Lysak.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Common dandelion (Taraxacum officinale) collecting locations and estimated genome sizes.
Rights and permissions
About this article
Cite this article
Iaffaldano, B.J., Zhang, Y., Cardina, J. et al. Genome size variation among common dandelion accessions informs their mode of reproduction and suggests the absence of sexual diploids in North America. Plant Syst Evol 303, 719–725 (2017). https://doi.org/10.1007/s00606-017-1402-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1402-2