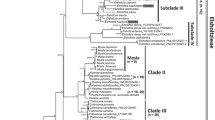
Abstract
Reconstruction of molecular phylogenies is an important step towards understanding the evolutionary history of island plant radiations. The New Zealand forget-me-nots (Myosotis, Boraginaceae) comprise a lineage of over 40 closely related but morphologically and ecologically diverse species whose evolutionary history and taxonomy are unclear. Myosotis is a high priority for systematic research in New Zealand because a high proportion of these species are threatened, and many have restricted geographic ranges and occupy very specific habitats. Here, we investigated the relationships and age of Southern Hemisphere forget-me-nots by performing phylogenetic, molecular dating, and other analyses of DNA sequence datasets from representatives of nearly all described species. To this end, we used both chloroplast (atpI–atpH + rps16–trnQ) and nuclear ribosomal (ITS + ETS) DNA sequences, as well as amplified fragment length polymorphisms (AFLPs). Our analyses showed that genus Myosotis likely arose in the Northern Hemisphere during the Miocene with the ancestor of the Southern Hemisphere lineage arising in the Pleistocene and radiating shortly thereafter. The Southern Hemisphere Myosotis species have very low levels of genetic divergence and their relationships are largely unresolved, likely due to a combination of recent radiation, hybridization, and incomplete lineage sorting. Our results are compared to those of similar studies on other New Zealand species radiations, and implications for ongoing and future Myosotis taxonomic and evolutionary research are discussed.




Similar content being viewed by others
References
Albach DC, Meudt HM (2010) Phylogeny of Veronica in the Southern and Northern Hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Molec Phylogenet Evol 54:457–471
Bacon CD, Allan GJ, Zimmer EA, Wagner WL (2011) Genome scans reveal high levels of gene flow in Hawaiian Pittosporum. Taxon 60:733–741
Baldwin BG, Markos S (1998) Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae). Molec Phylogenet Evol 10:449–463
Beuzenberg EJ, Hair JB (1983) Contributions to a chromosome atlas of the New Zealand flora—25 miscellaneous species. New Zealand J Bot 21:13–20
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537
Brandon AM (2001) Breeding systems and rarity in New Zealand Myosotis. Dissertation, Massey University, Palmerston North, New Zealand
Breitwieser I, Brownsey PJ, Garnock-Jones PJ, Perrie LR, Wilton A (2012) Phylum Tracheophyta, vascular plants. In: Gordon D (ed) The New Zealand inventory of biodiversity: a species 2000 symposium review. University of Canterbury Press, Christchurch, pp 411–459
Cohen J, Davis J (2009) Nomenclatural changes in Lithospermum (Boraginaceae) and related taxa following a reassessment of phylogenetic relationships. Brittonia 61:101–111
Cohen J (2010) Continuous characters in phylogenetic analyses: patterns of corolla tube length evolution in Lithospermum L. (Boraginaceae). Biol J Linn Soc 107:442–457
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Meth 9:772
de Candolle AP (1846) Prodromus systematis naturalis regni vegetabilis. Treuttel et Wurtz, Paris, France
de Lange PJ, Murray BG (2002) Contributions to a chromosome atlas of the New Zealand flora—37 miscellaneous families. New Zealand J Bot 40:1–23
de Lange PJ, Rolfe J (2010) New Zealand Indigenous Vascular Plant Checklist. New Zealand Plant Conservation Network, Wellington, New Zealand
de Lange PJ, Norton DA, Courtney SP, Heenan PB, Barkla JW, Cameron EK, Hitchmough R, Townsend AJ (2009) Threatened and uncommon plants of New Zealand (2008 revision). New Zealand J Bot 47:61–96
de Lange PJ, Rolfe JR, Champion PD, Courtney SP, Heenan PB, Barkla JW, Cameron EW, Norton DA, Hitchmough RA (2013) Conservation status of New Zealand indigenous vascular plants, 2012 New Zealand threat classification series 3. Department of Conservation, Wellington, New Zealand
Doyle JF, Dickson EE (1987) Preservation of plant samples for DNA restriction endonuclease analyses. Taxon 36:715–722
Druce AP (1993) Indigenous vascular plants of New Zealand, 9th revision. Landcare Research, Lower Hutt
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:699–710
Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A (2007) Geneious v 5.3.6 < http://www.geneiouscom/>
Ferrero V, Arroyo J, Vargas P, Thompson JD, Navarro L (2009) Evolutionary transitions of style polymorphisms in Lithodora (Boraginaceae). Perspect Pl Ecol Evol Syst 11:111–125
Fisher FJF (1965) The Alpine Ranunculi of New Zealand. New Zealand Department of Scientific and Industrial Research Bulletin 165:1–192
Ford KA, Ward JM, Smissen RD, Wagstaff SJ, Breitwieser I (2007) Phylogeny and biogeography of Craspedia (Asteraceae: Gnaphalieae) based on ITS, ETS and psbA-trnH sequence data. Taxon 56:783–794
Gabel ML (1987) Fossil Lithospermum (Boraginaceae) from the Tertiary of South Dakota. Amer J Bot 74:1690–1693
Gernhard T (2008a) The conditioned reconstructed process. J Theor Biol 253:769–778
Gernhard T (2008b) New analytic results for speciation times in neutral models. Bull Math Biol 70:1082–1097
Given DR (1981) Rare and endangered plants of New Zealand. Christchurch, AH and A W Reed Ltd
Grau J, Schwab A (1982) Mikromerkmale der Blüte zur Gliederung der Gattung Myosotis. Mitt Bot Staatssamml München 18:8–58
Guindon S, Gascuel O (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol 52:696–704
Gürke (1893) Boraginaceae In: Engler, Prantl (eds) Nat Pflanzenfam, pp 71–131
Gussarova G, Popp M, Vitek E, Brochmann C (2008) Molecular phylogeny and biogeography of the bipolar Euphrasia (Orobanchaceae): Recent radiations in and old genus. Molec Phylogenet Evol 48:444–460
Hasenstab-Lehman KE, Simpson MG (2012) Cat’s eyes and popcorn flowers: Phylogenetic systematics of the genus Cryptantha s.l. (Boraginaceae). Syst Bot 37:738–757
Heenan PB, McGlone MS (2013) Evolution of New Zealand alpine and open-habitat plant species during the late Cenozoic. New Zealand J Ecol 37:105–113
Hilger HH, Selvi F, Papini A, Bigazzi M (2004) Molecular systematics of Boraginaceae tribe Boragineae based on ITS1 and trnL sequences, with special reference to Anchusa s.l. Ann Bot (Oxford) 94:201–212
Hollis CJ, Beu AG, Crampton JS, Crundwell MP, Morgans HEG, Raine JI, Jones CM, Boyes AF (2010) Calibration of the New Zealand Cretaceous-Cenzoic timescale to GTS2004. GNS Sci Rep 43:1–20
Ho SYW, Phillips MJ (2009) Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol 58:367–380
Howarth DG, Baum DA (2002) Phylogenetic utility of a nuclear intron from nitrate reductase for the study of closely related plant species. Molec Phylogenet Evol 23:525–528
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molec Biol Evol 23:254–267
Jesson LK (2007) Ecological correlates of diversification in New Zealand angiosperm lineages. New Zealand J Bot 45:35–51
Joly S, Heenan PB, Lockhart PJ (2009) A Pleistocene inter-tribal allopolyploidization event precedes the species radiation of Pachycladon (Brassicaceae) in New Zealand. Molec Phylogenet Evol 51:365–372
Joly S, Heenan PB, Lockhart PJ (2014) Species radiation by niche shifts in New Zealand’s rockcresses (Pachycladon, Brassicaceae). Syst Biol 63:192–202
Khoshsokhan Mozaffar M, Kazempour Osaloo S, Oskoueiyan R, Naderi Saffar K, Amirahmadi A (2013) Tribe Eritrichieae (Boraginaceae s. str.) in West Asia: a molecular phylogenetic perspective. Pl Syst Evol 299:197–208
Kolarčik V, Zozomová-Lihová J, Mártonfi P (2010) Systematics and evolutionary history of the Asterotricha group of the genus Onosma (Boraginaceae) in central and southern Europe inferred from AFLP and nrDNA ITS data. Pl Syst Evol 290:21–45
Långström E, Chase MW (2002) Tribes of Boraginoideae (Boraginaceae) and placement of Antiphytum, Echiochilon, Ogastemma and Sericostoma: a phylogenetic analysis based on atpB plastid DNA sequence data. Pl Syst Evol 234:137–153
Långström E, Oxelman B (2003) Phylogeny of Echiochilon (Echiochileae, Boraginaceae) based on ITS sequences and morphology. Taxon 52:725–735
Lee WG, Tanentzap AJ, Heenan PB (2012) Plant radiation history affects community assembly: evidence from the New Zealand alpine. Biol Lett. doi:10.1098/rsbl20111210
Lehnebach CA (2008) Phylogenetic affinities, species delimitation and adaptive radiation of New Zealand Ranunculus. Dissertation, Massey University, Palmerston North, New Zealand
Lehnebach CA (2012) Two new species of forget-me-nots (Myosotis, Boraginaceae) from New Zealand. PhytoKeys 16:53–64. doi:10.3897/phytokeys163602
Levin RA, Blanton J, Miller JS (2009) Phylogenetic utility of nuclear nitrate reductase: A multi-locus comparison of nuclear and chloroplast sequence data for inference of relationships among American Lycieae (Solanaceae). Molec Phylogenet Evol 50:608–617
Li M, Wunder J, Bissoli G, Scarponi E, Gazzani S, Barbaro E, Saedler H, Varotto C (2008) Development of COS genes as universally amplifiable markers for phylogenetic reconstructions of closely related plant species. Cladistics 24:727–745
Lockhart PJ, McLenachan PA, Havell D, Glenny D, Huson D, Jensen U (2001) Phylogeny, radiation, and transoceanic dispersal of New Zealand alpine buttercups: molecular evidence under split decomposition. Ann Missouri Bot Gard 88:458–477
Mabberley DJ (2008) Mabberley’s Plant-book, 3rd edn. Cambridge University Press, Cambridge
Mandáková T, Heenan PB, Lysak MA (2010) Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evol Biol 10:367
Mansion G, Selvi F, Guggisberg A, Conti E (2009) Origin of Mediterranean insular endemics in the Boraginales: Integrative evidence from molecular dating and ancestral area reconstruction. J Biogeogr 36:1282–1296
Meudt HM (2006) A monograph of the genus Ourisia Comm. ex Juss. (Plantaginaceae). Syst Bot Monogr 77:1–188
Meudt HM (2008) Taxonomic revision of Australasian snow hebes (Veronica, Plantaginaceae). Austral Syst Bot 21:387–421
Meudt HM (2011) Amplified fragment length polymorphism data reveal a history of auto- and allopolyploidy in New Zealand endemic species of Plantago (Plantaginaceae): new perspectives on a taxonomically challenging group. Int J Pl Sci 172:220–237
Meudt HM (2012) A taxonomic revision of native New Zealand Plantago (Plantaginaceae). New Zealand J Bot 50:101–178
Meudt HM, Bayly MJ (2008) Phylogeographic patterns in the Australasian genus Chionohebe (Veronica s.l., Plantaginaceae) based on AFLP and chloroplast DNA sequences. Molec Phylogenet Evol 47:319–338
Meudt HM, Lockhart PJ, Bryant D (2009) Species delimitation and phylogeny of a New Zealand plant species radiation. BMC Evol Biol 9:111
Meudt HM, Prebble JM, Stanley RC, Thorsen MJ (2013) Morphological and amplified fragment length polymorphism (AFLP) data show that New Zealand endemic Myosotis petiolata (Boraginaceae) comprises three rare and threatened species. Austral Syst Bot 26:210–232
Mitchell AD, Heenan PB, Murray BG, Molloy BPJ, de Lange PJ (2009) Evolution of the south-western Pacific genus Melicytus (Violaceae): evidence from DNA sequence data, cytology and sex expression. Austral Syst Bot 22:143–157
Moore LB (1961) Boraginaceae. In: Allan HH (ed) Flora of New Zealand. PD Hasselberg, Government printer Wellington, pp 806–833
Moore MJ, Jansen RK (2006) Molecular evidence for the age, origin, and evolutionary history of the American desert plant genus Tiquilia (Boraginaceae). Molec Phylogenet Evol 39:668–687
Moore LB, Simpson JA (1973) A new Myosotis from north-west Nelson. New Zealand J Bot 11:163–170
Murray B, de Lange PJ (2013) Contributions to a chromosome atlas of the New Zealand flora – 40 miscellaneous counts for 36 families. New Zealand J Bot 51:31–60
Nazaire M, Wang XQ, Hufford L (2014) Geographic origins and patterns of radiation of Mertensia (Boraginaceae). Amer J Bot 101:104–118
Nazaire M, Hufford L (2012) A broad phylogenetic analysis of Boraginaceae: implications for the relationships of Mertensia. Syst Bot 37:758–783
Peralta IE, Spooner DM (2001) Granule-bound starch synthase (GBSSI) gene phylogeny of wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon). Amer J Bot 88:1888–1902
Pirie MD, Lloyd KM, Lee WG, Linder HP (2010) Diversification of Chionochloa (Poaceae) and biogeography of the New Zealand Southern Alps. J Biogeogr 37:379–392
Prebble JM, Meudt HM, Garnock-Jones PJ (2012) An expanded molecular phylogeny of the southern bluebells (Wahlenbergia, Campanulaceae) from Australia and New Zealand. Austral Syst Bot 25:11–30
Rambaut A, Drummond AJ (2007) Tracer v16. Available from http://www.beastbioedacuk/Tracer
Rambaut A, Drummond AJ (2010) LogCombiner v1 54 MCMC Output Combiner. University of Edinburgh, Institute of Evolutionary Biology
Rambaut A, Drummond AJ (2013) TreeAnnotator v1 70. University of Edinburgh, Institute of Evolutionary Biology
Robertson AW (1989) Evolution and pollination of New Zealand Myosotis (Boraginaceae). Dissertation, University of Canterbury, Christchurch
Robertson AW (1992) The relationship between floral display size, pollen carryover and geitonogamy in Myosotis colensoi (Kirk) Macbride (Boraginaceae). Biol J Linn Soc 46:333–349
Robertson AW, MacNair MR (1995) The effects of floral display size on pollinator service to individual flowers of Myosotis and Mimulus. Oikos 72:106–114
Rogers G, Walker S, Tubbs M, Henderson J (2002) Ecology and conservation status of three “spring annual” herbs in dryland ecosystems of New Zealand. New Zealand J Bot 40:649–669
Segal R (1966) Taxonomic study of the fossil species of the genus Cryptantha (Boraginaceae). Southwest Nat 11:205–210
Schmidt-Lebuhn AN, Kessler M, Kumar M (2006) Promiscuity in the Andes: species relationships in Polylepis (Rosaceae, Sanguisorbeae) based on AFLP and morphology. Syst Bot 31:547–559
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer J Bot 92:142–166
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Amer J Bot 94:275–288
Skinner MF, Johnson FW (1984) Tertiary stratigraphy and the Frick collection of fossil vertebrates from north-central Nebraska. Bull Amer Museum Nat Hist 178:215–368
Smissen RD, Galbany-Casals M, Breitwieser I (2011) Ancient allopolyploidy in the everlasting daisies (Asteraceae: Gnaphalieae): Complex relationships among extant clades. Taxon 60:649–662
Smissen RD, Morse CW, Prada D, Ramón-Laca A, Richardson SJ (2012) Characterisation of seven polymorphic microsatellites for Nothofagus subgenus Fuscospora from New Zealand. New Zealand J Bot 50:227–231
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stanley RJ, Dickinson KJM, Mark AF (1998) Demography of a rare endemic Myosotis: boom and bust in the high-alpine zone of southern New Zealand. Arctic Alpine Res 30:227–240
Steele PR, Guisinger-Bellian M, Linder CR, Jansen RK (2008) Phylogenetic utility of 141 low-copy nuclear regions in taxa at different taxonomic levels in two distantly related families of rosids. Molec Phylogenet Evol 48:1013–1026
Valdés B (2004) The Euro + Med treatment of Boraginaceae. Willdenowia 34:59–61
Wagstaff SJ, Bayly MJ, Garnock-Jones PJ, Albach DC (2002) Classification, origin and diversification of the New Zealand hebes (Scrophulariaceae). Ann Missouri Bot Gard 89:38–63
Wagstaff SJ, Breitwieser I (2004) Phylogeny and classification of Brachyglottis (Senecioneae, Asteraceae): An example of a rapid species radiation in New Zealand. Syst Bot 29:1003–1010
Wagstaff SJ, Garnock-Jones PJ (2000) Patterns and diversification of Chionohebe and Parahebe (Scrophulariaceae) inferred from ITS sequences. New Zealand J Bot 38:389–407
Webb CJ, Sykes WR, Garnock-Jones PJ (1988) Flora of New Zealand, vol IV. DSIR, Christchurch
Weeks A, Simpson BB (2004) Molecular genetic evidence for interspecific hybridization among endemic Hispaniolan Bursera (Burseraceae). Amer J Bot 91:976–984
Weigend M, Gottschling M, Selvi F, Hilger HH (2009) Marbleseeds are gromwells—systematics and evolution of Lithospermum and allies (Boraginaceae tribe Lithospermeae) based on molecular and morphological data. Molec Phylogenet Evol 52:755–768
Weigend M, Gottschling M, Selvi F, Hilger HH (2010) Fossil and extant western hemisphere Boragineae, and the polyphyly of “Trigonotideae” Riedl. (Boraginaceae: Boraginoideae). Syst Bot 35:409–419
Weigend M, Luebert F, Selvi F, Brokamp G, Hilger HH (2013) Multiple origins for hound’s tongues (Cynoglossum L.) and navel seeds (Omphalodes Mill.)–The phylogeny of the borage family (Boraginaceae s. str.). Molec Phylogenet Evol 68:604–618
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis, MA, Gelfand, DH, Sninsky, JJ, and White, TJ, (eds), PCR Protocols: a guide to methods and applications. Academic Press, Inc., pp 315–322
Winkworth RC, Grau J, Robertson AW, Lockhart PJ (2002) The origins and evolution of the genus Myosotis L. (Boraginaceae). Molec Phylogenet Evol 24:180–193
Acknowledgments
We would like to thank Pat Brownsey and Ilse Breitwieser for their support of this project, staff from AK, CHR and WELT, the Department of Conservation (DOC) for permission and help to collect samples on DOC land, the many colleagues who helped with field work and sample collection (especially John Barkla, Andrea Brandon, Fiona Cameron, Hamish Carson, Bill Clarkson, Shannel Courtney, Pat Enright, Phil Garnock-Jones, David Glenny, Ant Kusabs, Alan Lee, Mike Lusk, Viv McGlynn, Brian Rance, Nick Singers, Jorge Santos, Barry Sneddon, Bec Stanley, Mike Thorsen, Steve Wagstaff, and Andreas Zeller), Dirk Albach, Dick Olmstead, and Matt Renner for sending Boraginaceae outgroup tissue and DNA, Peter Ritchie for use of the Molecular Ecology lab at Victoria University of Wellington to generate the DNA sequence and AFLP data, Simon Pfanzelt for key discussions and unwavering help running BEAST analyses, Phil Garnock-Jones for commenting on an earlier version of the manuscript, and two anonymous reviewers for their highly constructive criticisms during the review process. We acknowledge use of information contained in the New Zealand Fossil Record File. This research was supported by Core funding for Crown Research Institutes from the Ministry of Business, Innovation and Employment’s Science and Innovation Group.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Hervé Sauquet.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic Supplementary Material
606_2014_1166_MOESM1_ESM.pdf
Online Resource 1 Table showing voucher details including GenBank accession numbers. ITS sequences downloaded from GenBank include a reference; all other sequences were newly sequenced for this study. Author names are given for species, subspecies and varieties not listed in Table 1 (PDF 150 kb)
606_2014_1166_MOESM2_ESM.pdf
Online Resource 2 RAxML phylogram of combined nuclear (ITS + ETS) DNA sequence dataset; scale units are substitutions per site. Individual names are genus + species epithet + Genbank number for the atpI–atpH sequence (or if no atpI–atpH sequence, then Genbank number for the ITS sequence). Numbers above branches are bootstrap values from 1000 replicates. L, C and M indicate the location of the calibration points based on Lithospermum, Cryptantha and New Zealand Myosotis fossils respectively (note these are shown here for illustrative purposes only as they were used in the BEAST analyses not in this RAxML anlaysis; see text for details). Voucher information for all individuals is given in Online Resource 1 (PDF 978 kb)
606_2014_1166_MOESM3_ESM.pdf
Online Resource 3 RAxML phylogram of combined chloroplast DNA (rps16-trnQ+atpI–atpH intergenic spacer regions) dataset; scale units are substitutions per site. Individual names are genus + species epithet + Genbank number for the atpI–atpH sequence (or if no atpI–atpH sequence, then Genbank number for the ITS sequence). Numbers above branches are bootstrap values from 1000 replicates. Voucher information for all individuals is given in Online Resource 1 (PDF 526 kb)
606_2014_1166_MOESM4_ESM.pdf
Online Resource 4 BEAST chronogram of combined nuclear (ITS + ETS) DNA sequence dataset using Lithospermum (L) and Cryptantha (C) fossil calibration points with uniform priors (the Myosotis fossil calibration point [M] was not used in this analysis; see text for details). The scale line is in millions of years before present. Individual names are genus + species epithet + Genbank number for the atpI–atpH sequence (or if no atpI–atpH sequence, then Genbank number for the ITS sequence). Dark circles signify branches with posterior probability values >0.95. Voucher information for all individuals is given in Online Resource 1 (PDF 49 kb)
Rights and permissions
About this article
Cite this article
Meudt, H.M., Prebble, J.M. & Lehnebach, C.A. Native New Zealand forget-me-nots (Myosotis, Boraginaceae) comprise a Pleistocene species radiation with very low genetic divergence. Plant Syst Evol 301, 1455–1471 (2015). https://doi.org/10.1007/s00606-014-1166-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-014-1166-x