Abstract
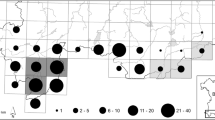
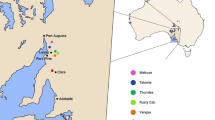
Sloe (Prunus spinosa L.) is a shrub native to Europe. In Germany, 50–80 % of all planted sloe is imported. Little is known about the genetic diversity patterns within and between German sloe populations. Thus, a debate arose how to avoid risks for nature and landscape by planting potentially maladapted material. The main objectives of our study are to analyse the genetic differentiation pattern of sloe populations in Germany, to identify geographic/genetic structures and to evaluate their potential for tracing reproductive material. 17 natural populations from Germany and 1 from Italy and Hungary were investigated by Amplified Fragment Length Polymorphisms (AFLP) and PCR–RFLP techniques. The AMOVA analyses based on AFLPs for all populations and for the German populations only result in equally high differentiation values of ΦPT = 15 % of molecular variance between populations. The analysis of cpDNA PCR–RFLPs resulted in 24 haplotypes with 30 % showing genetic variation between populations. Overall values of genetic variability over all loci and populations are: Na = 0.832, Ne = 1.114 and He = 0.072. Mantel tests for AFLPs and cpDNA haplotypes reveal no association between geographic and genetic distances between populations as a result of a lack of differentiation between German populations and those from southern and southeastern Europe. Weak geographic/genetic patterns were observed on a large scale. However, these concern the German populations only. Our results indicate that vegetative regeneration in combination with founder effects may influence the level of differentiation between populations. Populations with a large amount of vegetative propagation are more differentiated from other populations than those populations which exhibit less vegetative regeneration. The assignment of reproductive material (i.e. plant material) to potential source populations resulted in high values of correct allocations. Hence, such methods can be applied to trace reproductive material of unknown origin.






Similar content being viewed by others
References
Aguinagalde I, Hampe A, Mohanty A, Martin JP, Duminil J, Petit RJ (2005) Effects of life history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. J Biogeogr 32:329–339
Bergmann F (1991) Causes and consequences of species-specific genetic variation patterns n european forest tree species. Examples with norway spruce and silver fir. In: Müller-Starck G, Ziehe M (eds) Genetic variation in European populations of forest trees. Sauerländers, Frankfurt am Main, pp 192–205
Deforce K, Bastiaens J, Van Neer W, Ervynck A, Lentacker A, Sergant J, Crombe P (2012) Wood charcoal and seeds as indicators for animal husbandry in a wetland site during the late Mesolithic–early Neolithic transition period (Swifterbant culture, ca. 4600–4000 BC) in NW Belgium. Vegetation Hist Archaeobotany 22:51–60. doi:10.1007/s00334-012-0354-2
Dorken ME, Eckert CG (2001) Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol 89:339–350
Duchesne P, Bernatchez L (2002) AFLPOP: a computer program for simulated and real population allocation, based on AFLP data. Mol Ecol Notes 2:380–383. doi:10.1046/j.1471-8278.2002.00251.x
Duminil J, Fineschi S, Hampe A, Jordano P, Salvini D, Vendramin GG, Petit RJ (2007) Can population genetic structure be predicted from life-history traits? Am Nat 169(5):662–672
Eimert K, Rückert FE, Schröder M-B (2012) Genetic diversity within and between seedstock populations of several German autochthonous provenances and conventionally propagated nursery material of blackthorn (Prunus spinosa L.). Plant Syst Evol 298:609–618
Eriksson O (1993) Dynamics of genets in clonal plants. Trends Ecol Evol 8:313–316
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics 131:479–491
Fronia R (2009) Prüfung der Identität und Variabilität gebeietsheimischer und gebietsfremder Herkünfte von Prunus spinosa L. und Cornus sanguinea L. zur Verwendung in der freien Land-schaft. Der Andere Verlag, Tönning, Lübeck, Marburg
Gailing O, von Wühlisch G (2004) Nuclear markers (AFLPs) and chloroplast microsatellites differ between Fagus sylvatica and F. orientalis. Silvae Genetica 53:105–110
Gillet EM (2010) GSED 3.0—Genetic Structures from Electrophoresis Data. http://www.uni-goettingen.de/de/67064.html
Gregorius H-R, Roberds JH (1986) Measurement of genetical differentiation among subpopulations. Theor Appl Genet 71, 826–834. http://dx.doi.org/10.1007/BF00276425
Guitán J, Guitán P, Sánchez JM (1993) Reproductive biology of two Prunus species (Rosaceae) in the northwest Iberian Peninsula. Pl Syst Evol 185:153–165
Hanelt P (1997) European wild relatives of Prunus fruit crops. In: Valdes B, Heywood VH, Raimondo FM, Zohary D (eds) Bocconea-7, Proceedings of the Workshop on “Conservation of the wild relatives of European cultivated plants”. Bocconea 7, Palermo, Italy, pp 401–408
Hebel I, Haas R, Dounavi A (2006) Genetic variation of common ash (Fraxinus excelsior L.) populations from provenance regions in southern Germany by using nuclear and chloroplast microsatellites. Silvae Genet 55:38–44
Hosius B, Leinemann L, Konnert M, Bergmann F (2006) Genetic aspects of forestry in the Central Europe. Eur J Forest Res 125:407–417
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss Buchloe dactyloides (Nutt.) Engelm. Theor Appl Genet 86:927–934
Kowarik I, Seitz B (2003) Perspektiven für die Verwendung gebietseigener (“autochthoner”) Gehölze. Neobiota 2:3–26
Kremer A, Kleinschmit J, Cottrell J, Cundall EP, Deans JD, Ducousso A, König AO, Lowe AJ, Munro RC, Petit RJ, Stephan BR (2002) Is there a correlation between chloroplastic and nuclear divergence, or what are the roles of history and selection on genetic diversity in European oaks? For Ecol Manag 156:75–87
Leinemann L (2000) Inheritance analysis of isozyme phenotypes in tetraploid species using single plant progenies. An example in sloe (Prunus spinosa L.). Forest Genetics 7(3):205–209
Kleinschmit J, Leinemann, L and Hosius B (2008) Gene Conservation through seed orchards—a case study of Prunus spinosa L. In: Seed Orchard Conference Dag Lindgren (ed), Umea, Sweden pp 115–126
Leinemann L, Bendixen K, Kownatzki D, Hattemer HH, Liepe K, Stenger G (2002) Genetische Untersuchungen an Land-schaftsgehölten im Hinblick auf die Erzeugung und Zertifizierung von Vermehrungsgut. Allg Forst u J-Ztg 173:145–152
Leinemann L, Steiner W, Hosius B, Kleinschmit J (2010) Klonale Reproduktion in naturnahen Vorkommen der Schlehe (Prunus spinosa L.). Forstarchiv 81. Heft 4:165–169
Leinemann L, Steiner W, Hosius B, Kuchma O, Arenhövel W, Fussi B, Haase B, Kätzel R, Rogge M, Finkeldey R (2013) Genetic variation of chloroplast and nuclear markers in natural populations of hazelnut (Corylus avellana L.) in Germany. Plant Syst Evol 299:369–378. doi:10.1007/s00606-012-0727-0
Liepelt S, Chedaddi R, de Beaulieu J-L, Gomory D, Hussendörfer E, Konnert M, Litt T, Longauer R, Terhürne-Berson R, Ziegenhagen B (2009) Postglacial range expansion and genetic imprints in Abies alba Mill—a synthesis based on palaeobotany and genetics. Rev Palaeoontology Palaeobotany 153:139–149
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Ann Rev Ecol Syst 15:65–96
Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Lataowa M, Litt T, Paule L, Roure JM, Tantau I, van der Knaap WO, Petit RJ, de Beaulieu JL (2006) A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol 171:199–221
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Martin L, Jacomet S, Thiebault S (2008) Plant economy during the Neolithic in a mountain context: the case of ′′Le Chenet des Pierres′′ in the French Alps (Bozel-Savoie, France). Vegetation Hist Archaeobotany 17:113–122
Michalakis Y, Excoffier L (1996) A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142:1061–1064
Mohanty A, Martín JP, Aguinagalde I (2000) Chloroplast DNA diversity within and among populations of the allotetraploid Prunus spinosa L. Theor Appl Genet 100:1304–1310
Mohanty A, Martín JP, Aguinagalde I (2002) Population genetic analysis of European Prunus spinosa (Rosaceae) using chloroplast DNA markers. Am J Bot 89:1223–1228
Moran PAP (1950) Notes on continuous stochastic phenomena. Biometrika 37:17–23
Nathan R, Perry G, Cronin JT, Strand AE, Cain ML (2003) Methods for estimating long-distance dispersal. Oikos 103:261–273
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Peakall R, Smouse PE, Huff DR (1995) Evolutionary implications of allozyme and RAPD variation in diploid populations of dioecious buffalograss Buchloe dactyloides. Mol Ecol 4:135–147
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57:1182–1195
Petit R, Csaikl U, Bordács S, Burg K, Coart E, Cottrell J, van Dam B, Deans D, Dumolin-Lapègue S, Fineschi S, Finkeldey R, Gillies A, Glaz I, Goicoechea PG, Jensen JS, König AO, Lowe AJ, Madsen SF, Mátyás G, Munro RC, Olalde M, Pemonge M-H, Popescu F, Slade D, Tabbener H, Taurchini D, de Vries SGM, Ziegenhagen B, Kremer A (2002) Chloroplast DNA variation in European white oaks. Phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manag 156:5–26
Petit R, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701. doi:10.1111/j.1365-294X.2004.02410
Schmidt S (2003) Genetische Vielfalt und Vernetzung verschiedener Teilpopulationen von Coryllus avellana L. und Prunus spinosa L. an Wald- und Wegrändern des Sollings. Dissertation, Universität Göttingen
Scholz H, Scholz I (1995) Unterfamilie Prunoideae. In: Hegi G (ed) Illustrierte Flora von Mitteleuropa, 2. Aufl. Bd. IV, Teil 2B, Lfg. 6 & 7. Blackwell, Berlin, pp 446–510
Spethmann W (1995) In-situ/ex-situ-Erhaltung von heimischen Straucharten. Schriften zu Genetischen Ressourcen. ZADI Bonn 1:68–87
Spethmann W (2003) Wie können Saatguthandel und Baumschulen einen Beitrag zur Erhaltung der Biodiversität einheimischer Sträucher leisten? Neobiota 2:27–35
Tollefsrud MM, Sønstebø JH, Brochmann C, Johnsen Ø, Skrøppa T, Vendramin GG (2009) Combined analysis of nuclear and mitochondrial markers provide new insight into the genetic structure of North European Picea abies. Heredity 102:549–562
Vander Mijnsbrugge K, Depypere L, Chaerle P, Goetghebeur P, Breyne P (2013) Genetic and morphological variability among autochthonous Prunus spinosa populations in Flanders (northern part of Belgium): implications for seed sourcing. Plant Ecol Evol 146(2):193–202
Vieira J, Ram Santos, Habu T, Tao R, Vieira CP (2008) The Prunus Self-Incompatibility Locus (S locus) is seldom rearranged. J Heredity 99(6):657–660
Vos P, Hogers R, Bleeker M, Reijans M, van de lee T, Frijters A, Pot J, Peleman J, Kiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23(21):4407–4414
Yeboah GK, Woodel SR (1987) Flowering phenology, flower colour, and mode of reproduction of Prunus spinosa L. (blackthorn); Crataegus monogyna Jacq. (hawthorn); Rosa canina L. (dog rose); and Rubus fruticosus (bramble) in Oxfordshire. Engl Funct Ecol 1:261–268
Acknowledgments
We thank Olga Artes, Alexandra Lindner und Barbara Buchwinkler for their outstanding work in the lab. The kind and efficient support of Dr. Norbert Kowarsch (BLE, Referat 514, Projektträger Agrarforschung) during all phases of the project is highly appreciated. This work was financially supported by the German Federal Ministry of Food, Agriculture and Consumer Protection (BMELV) through the Federal Office for Agriculture and Food (BLE), grant number 2807BM029.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Leinemann, L., Kleinschmit, J., Fussi, B. et al. Genetic composition and differentiation of sloe (Prunus spinosa L.) populations in Germany with respect to the tracing of reproductive plant material. Plant Syst Evol 300, 2115–2125 (2014). https://doi.org/10.1007/s00606-014-1027-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-014-1027-7