Abstract
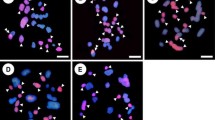
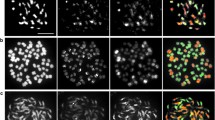
Genus Helianthus comprises diploid and polyploid species. An autoallopolyploid origin has been proposed for hexaploid species but the genomic relationships remain unclear. Mitotic and meiotic studies in annual Helianthus annuus (2n = 2x = 34) and perennial Helianthus resinosus (2n = 6x = 102) as well as the F1 hybrids between both species were carried out. Chromosome counting confirmed the hybrid origin of the latter plants and their tetraploid condition. Bivalents in hybrids ranged from 12 to 28 (\( \bar{x} \) = 20.8). Univalents, trivalents and quadrivalents were also observed. Meiotic products comprised dyads, triads and normal tetrads and pollen grains were heterogeneous in size. These observations suggest the occurrence of 2n pollen in addition to the expected n. Genomic in situ hybridization (GISH) of total H. annuus DNA on H. resinosus chromosomes rendered weak but uniform signals; similar hybridization pattern was observed using three other annual species. Hybridization with H. annuus probe performed on root tip cells of F1 H. annuus × H. resinosus hybrids revealed 17 chromosomes with a strong hybridization signal. GISH in hybrid meiocytes distinguished chromosomes from parental species and revealed autosyndetic pairing of H. resinosus chromosomes, allosyndetic pairing in bivalents, trivalents and quadrivalents, and the presence of univalents derived from parents, H. annuus and H. resinosus. Results obtained from classical and molecular cytogenetics do not support H. annuus as a direct ancestor of H. resinosus. The occurrence of allosyndetic pairing and the relatively high fertility of the F1 hybrids point to the possibility that useful genes could be transferred from H. resinosus to cultivate sunflower, although the effective rate of recombination has not been evaluated. GISH method proved effective to recognize parental chromosomes in H. annuus × H. resinosus progeny.



Similar content being viewed by others
Abbreviations
- CTAB:
-
Cetyltrimethylammonium bromide
- ETS:
-
External Transcribed Spacer
- FISH:
-
Fluorescence in situ hybridization
- GISH:
-
Genomic in situ hybridization
- ITS:
-
Internal Transcribed Spacer
References
Atlagic J (1996) Cytogenetic studies in hexaploid Helianthus species and their F1 hybrids with cultivated sunflower, H. annuus. Plant Breed 115:257–260
Atlagic J, Dozet B, Skoric D (1995) Meiosis and pollen grain viability in Helianthus mollis, Helianthus salicifolius, Helianthus maximiliani and their F1 hybrids with cultivated sunflower. Euphytica 81:259–263
Bennett ST, Kenton AY, Bennett MD (1992) Genomic in situ hybridization reveals the allopolyploid nature of Milium montianum (Gramineae). Chromosoma 101:420–424
Camadro EL, Saffarano SK, Espinillo JC, Castro M, Simon PW (2008) Cytological mechanisms of 2n pollen formation in the wild potato Solanum okadae and pollen-pistil relations with the cultivated potato, Solanum tuberosum. Genet Resour Crop Ev 55:471–477
Carrera A, Poverene M, Rodriguez R (2004) Isozyme and cytogenetic analysis in Helianthus resinosus Small. Proc Intern Sunflower Conf. Vol II, Fargo, pp 685–691
Cavallini A, Natali L, Zuccolo A, Giordani T, Jurman I, Ferrillo V, Vitacolonna N, Sarri V, Cattonaro F, Ceccarelli M, Cionini PG, Morgante M (2010) Analysis of transposons and repeat composition of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 120:491–508
Ceccarelli M, Sarri V, Natali L et al (2007) Characterization of the chromosome complement of Helianthus annuus by in situ hybridization of a tandemly repeated DNA sequence. Genome 50:429–434
Chandler JM (1991) Chromosome evolution in sunflower. In: Tsuchiya P, Gupta PK (eds) Chromosome engineering in plants: genetics, breeding, evolution. Part B. Elsevier, Amsterdam, pp 229–249
Chester M, Leitch AR, Soltis PS, Soltis DE (2010) Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridization). Genes 1:166–192
CIMMYT (2005) Laboratory Protocols: CIMMYT Applied Molecular Genetics Laboratory, 3rd edn. CIMMYT, Mexico, DF
Cuellar T, Belhassen E, Fernandez-Calvin B, Orellana J, Bella JL (1996) Chromosomal differentiation in Helianthus annuus var macrocarpus: heterochromatin characterization and rDNA location. Heredity 76:586–591
Echeverría MM, Salaberry MT, Rodríguez RH (2003) Characterization for agronomic use of cytoplasmic male-sterility in sunflower (Helianthus annuus L.) introduced from H. resinosus small. Plant Breed 122:357–361
Espinasse A, Foueillassarl J, Kimber G (1995) Cytogenetical analysis of hybrids between sunflower and four wild relatives. Euphytica 82:65–72
Feng J, Liu Z, Cai X, Jan Ch (2013) Toward a molecular cytogenetic map for cultivated sunflower (Helianthus annuus L.) by landed BAC/BIBAC clones. G3 (Bethesda) 3(1):31–40. doi:10.1534/g3.112.004846
Fernandez P, Paniego N, Lew S, Hopp HE, Heinz RA (2003) Differential representation of sunflower ESTs in enriched organ-specific cDNA libraries in a small scale sequencing project. BMC Genomics 4:40. doi:10.1186/1471-2164-4-40
Garayalde A, Poverene M, Cantamutto M, Carrera A (2011) Wild sunflower diversity in Argentina revealed by ISSR and SSR markers: an approach for conservation and breeding programmes. Ann Appl Biol 158:305–317
Giordani T, Natali L, Cavallini A (2003) Analysis of a dehydrin encoding gene and its phylogenetic utility in Helianthus. Theor Appl Genet 107:316–325
Heiser CB, Smith DM, Clevenger S, Martin WC (1969) The North American sunflowers (Helianthus). Mem Torrey Bot Club 22:1–218
Humphreys MW, Thomas HM, Morgan WG, Meredith MR, Harper JA, Thomas H, Zwierzykowski A, Ghesquiere M (1995) Discriminating the ancestral progenitors of hexaploid Festuca arundinacea using genomic in situ hybridization. Heredity 75:171–174
Jan CC, Seiler GJ (2007) Sunflower. In: Singh RJ (ed) Genetics Resources, Chromosome Engineering, and Crop Improvement, vol 4., Oilseed CropsCRC Press, Taylor and Francis Group, New York, pp 103–165
Kostoff D (1939) Autosynthesis and structure hybridity F1-hybrid H. tuberosus × H. annuus L. and their sequences. Genetica 21:285–300
Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson JJ, Grandbastien MA, Leitch AR (2007) Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol 175:756–763
Liu Z, Feng J, Jan CC (2009) Genomic in situ hybridization (GISH) as a tool to identify chromosomes of parental species in sunflower interspecific hybrids. Proc Nat Sunflower Assoc ARS USDA. http://hdl.handle.net/10113/34842
Liu Z, Cai X, Seiler GJ, Gulya TA, Rashid KY, Jan CC. 2013. Update on transferring Sclerotinia resistance genes from wild perennial Helianthus species into cultivated sunflower. National Sclerotinia Initiative 2013 Annual Meeting. http://www.ars.usda.gov/SP2UserFiles/ad_hoc/54000000WhiteMoldResearch/2013Meeting/2013%20National%20Sclerotinia%20Initiative%20Annual%20Meeting.pdf
Mok DWS, Peloquin SJ (1972) Three mechanisms of 2n pollen formation in diploid potatoes. Am Potato J 49:362–363
Mondolot-Cosson L, Andary C (1994) Resistance factors of a wild species of sunflower, Helianthus resinosus, to Sclerotinia sclerotiorum. Acta Hort (ISHS) 381:642–645
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34:401–437
Poggio L, Confalonieri V, Comas C, Cuadrado A, Jouve N, Naranjo CA (1999) Genomic in situ hybridization (GISH) of Tripsacum dactyloides and Zea mays ssp mays with B chromosomes. Genome 42:687–691
Prabakaran AJ, Sujatha M (2004) Interspecific hybrid of Helianthus annuus × H. simulans: characterization and utilization in improvement of cultivated sunflower (H. annuus L.). Euphytica 135:275–282
Ramanna MS, Jacobsen E (2003) Relevance of sexual polyploidization for crop improvement —A review. Euphytica 133:3–18
Rieseberg LH (1991) Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am J Bot 78:1218–1237
Rogers CE, Thompson TE, Seiler GJ (1982) Sunflower species of the United States. National Sunflower Association, Fargo
Sala CA, Echarte AM (1996) Cytological mechanism of 2n pollen formation in interspecific polyploid hybrids of Helianthus. In: International Sunflower Association (ed) 14th Intern Sunflower Conf, pp 73
Schilling EE (1997) Phylogenetic analysis of Helianthus (Asteraceae) based on chloroplast DNA restriction site data. Theor Appl Genet 94:925–933
Schilling EE, Heiser CB (1981) Infrageneric classification of Helianthus (Compositae). Taxon 30:393–403
Schilling EE, Linder CR, Noyes RD, Rieseberg LH (1998) Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Sys Bot 23:177–187
Sossey-Alaoui K, Serieys H, Tersac M, Lambert P, Schilling E, Griveau Y, Kaan F, Berville A (1998) Evidence for several genomes in Helianthus. Theor Appl Genet 97:422–430
Staton SE, Bakken BH, Blackman BK, Chapman MA, Kane NC, Tang S, Ungerer MC, Knapp SJ, Rieseberg LH, Burke JM (2012) The sunflower (Helianthus annuus L.) genome reflects a recent history of biased accumulation of transposable elements. Plant J 72:142–153
Sujatha M, Prabakaran AJ (2006) Ploidy manipulation and introgression of resistance to Alternaria helianthi from wild hexaploid Helianthus species to cultivated sunflower (H. annuus L.) aided by anther culture. Euphytica 152:201–215
Talia P, Greizerstein E, Díaz Quijano C et al (2010) Cytological characterization of sunflower by in situ hybridization using homologous rDNA sequences and a BAC clone containing highly represented repetitive. Genome 53:1–8
Taschetto OM, Pagliarini MS (2003) Occurrence of 2n and jumbo pollen in the Brazilian ginseng (Pfaffia glomerata and P. tuberosa). Euphytica 133:139–145
Thompson TE, Zimmerman DC, Rogers CE (1981) Wild Helianthus as a genetic resource. Field Crop Res 4:333–343
Timme RE, Simpson BB, Linder CR (2007) High-resolution phylogeny for Helianthus (Asteraceae) using the 18S-26S ribosomal DNA external transcribed spacer. Am J Bot 94:1837–1852
Veilleux R (1985) Diploid and polyploid gametes in crop plants: mechanisms of formation and utilization in plant breeding. Plant Breed Rev 3:253–288
Vukich M, Schulman AH, Giordani T, Natali L, Kalendar R, Cavallini (2009) Genetic variability in sunflower (Helianthus annuus L.) and in the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet 119:1027–1038
Acknowledgments
We are grateful to MSc. Raúl Rodríguez for his knowledge and help in getting the plant material. This research was supported by Grants ANPCYT-PICT 2286 and UNS-PGI 24A160.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miranda Zanetti, J., Greizerstein, E., Camadro, E. et al. Genomic relationships between hexaploid Helianthus resinosus and diploid Helianthus annuus (Asteraceae). Plant Syst Evol 300, 1071–1078 (2014). https://doi.org/10.1007/s00606-013-0945-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0945-0