Abstract
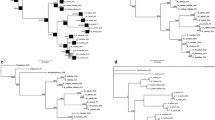
To establish relationships among the genera Blepharocalyx, Luma, and Myrceugenia, the phylogeny of tribe Myrteae was reconstructed based on secondary structures of the sequences of ITS (ITS1-5.8S-ITS2) and ETS regions from 93 taxa belonging to 29 genera. The DNA sequences were aligned according to their secondary structure and analysed with Bayesian inference (BI) using base substitution models for RNA, which can differentiate between the regions consisting of base pairs (helices) and unpaired bases (loops). The best-fit models were RNA7C for ITS and RNA7B for ETS, which differ in how they predict double substitutions and symmetry of the frequency of base pairs. The phylogenetic hypothesis was compared with results obtained using classical 4-state models, the results of maximum parsimony, and using sequence alignment without considering the secondary structure. Analyses show low support along the backbone of the consensus trees, those using substitution models of paired sites showing more highly supported derived clades than substitution models of independent sites. All tests were consistent and differed in the positioning of certain groups, but they also showed that Blepharocalyx, Luma, and Myrceugenia arise from three independent lineages that are not closely related. The substitution model for RNA shows that Luma is a monophyletic group and sister to the other genera of Myrteae, except for Myrtus communis. Myrceugenia appears as a monophyletic group, except for M. fernandeziana, which is related to Blepharocalyx, a genus that appears to be biphyletic.







Similar content being viewed by others
References
Berg O (1855–1856) Revisio Myrtacearum Americae. Linnaea 27:1–472
Berg O (1861) Mantissa II. Ad Revisionem Myrtacearum Americae. Linnaea 30:647–713
Biffin EE, Harrington MG, Crisp M, Craven L, Gadek P (2007) Structural partitioning, paired-sites models and evolution of the ITS transcript in Syzygium and Myrtaceae. Mol Phyl Evol 43:124–139
De Candolle AP (1828) Prodromus Systematis Naturalis Regni Vegetabilis Pars III. Sumptibus Sociorum Treuttel et Würtz, Paris
Fitch WM (1971) Toward defining the course of evolution, minimum change for a specific tree topology. Syst Zool 20:406–416
Goloboff P (1999). NONA (NO NAME), v.2. published by the author. Tucumán
Gowri V, Jow H (2006) PHASE: a software package for phylogenetics and sequence evolution. Version 2.0. University of Manchester
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Harpke D, Peterson A (2008) 5.8S motif for the identification of pseudogenic ITS regions. Botany 86:300–305
Harrington M, Biffin EE, Gadek PA (2009) Comparative study of the evolution of nuclear ribosomal spacers incorporating secondary structure analyses within Dodonaeoideae, Hippocastanoideae and Xanthoceroideae (Sapindaceae). Mol Phyl Evol 50:364–375
Hudelot C, Gowri-Shankar V, Jow H, Rattray M, Higgs P (2003) RNA-based phylogenetic methods: application to mammalian mitochondrial RNA sequences. Mol Phyl Evol 28:241–252
Jobes DV, Thien LB (1997) A conserved motif in the 5.8S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. Plant Mol Biol Rep 15:326–334
Jow H, Hudelot C, Rattray M, Higgs P (2002) Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Mol Biol Evol 19:1591–1601
Kausel E (1948) Notas mirtológicas. Lilloa 13:125–149
Kelchner SA (2000) The evolution of noncoding chloroplast DNA and its application in plant systematics. Ann Mo Bot Gard 87:482–498
Landrum LR (1981a) A monograph of the genus Myrceugenia (Myrtaceae). Flora Neotropica Monogr 29:1–135
Landrum LR (1981b) The phylogeny and geography of Myrceugenia (Myrtaceae). Brittonia 33:105–129
Landrum LR (1986) Campomanesia, Pimenta, Blepharocalyx, Legrandia, Acca, Myrrhinium, and Luma (Myrtaceae). Flora Neotropica Monogr 45:1–179
Landrum LR, Stevenson D (1986) Variability of embryos in subtribe Myrtinae (Myrtaceae). Syst Bot 11:155–162
Lee CY, Lee JC (1992) Structure analysis of the 5′ external transcribed spacer of the precursor ribosomal RNA from Saccharomyces cerevisiae. J Mol Biol 228:827–838
Legrand D (1957) Representantes neotropicales del género Myrceugenia. Darwiniana 11:293–365
Liston A, Robinson WA, Oliphant JM, Álvarez-B ER (1996) Length variation in the nuclear ribosomal internal transcribed spacer region of non-flowering seed plants. Syst Bot 21:109–120
Liu JS, Schardl CL (1994) A conserved sequence in internal transcribed spacer 1 of plant nuclear rRNA genes. Pl Mol Biol 21:109–120
Lucas E, Belsham SR, Lughadha EM, Orlovich DA, Sakuragui CM, Chase MW, Wilson PG (2005) Phylogenetic patterns in the fleshy-fruited Myrtaceae—preliminary molecular evidence. Pl Syst Evol 251:35–51
Lucas E, Harris S, Mazine F, Belsham SR, Lughadha EM, Telford A, Gasson P, Chase MW (2007) Suprageneric phylogenetics of Myrteae, the genetically richest tribe in Myrtaceae (Myrtales). Taxon 56:1105–1128
Mathews D, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288:911–940
McVaugh R (1968) The genera of American Myrtaceae, an interim report. Taxon 17:354–418
Mullineux T, Hausner G (2009) Evolution of rDNA ITS1 and ITS2 sequences and RNA secondary structures within members of the fungal genera Grosmannia and Leptographium. Fungal Genet Biol 46:855–867
Murillo-A J, Ruiz E, Landrum LR, Stuessy TF, Barfuss MHJ (2012) Phylogenetic relationships in Myrceugenia (Myrtaceae) based on plastid and nuclear DNA sequences. Mol Phyl Evol 62:764–776
Murillo-A J, Ruiz E (2011) Revalidación de Nothomyrcia (Myrtaceae), un género endémico del Archipiélago de Juan Fernández. Gayana Bot 68:129–134
Nixon KC (1999–2002) WinClada ver. 1.0000 Published by the author, Ithaca
Posada D, Crandall K (1998) Model test: testing the model of DNA substitution. Bioinformatics 14:817–818
Proença CE, Lughadha EM, Lucas E, Woodgyer EM (2006) Algrizea (Myrteae, Myrtaceae): a new genus from the Highlands of Brazil. Syst Bot 31:320–326
Rambaut A, Drummond AJ (2007) Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer
Savill NJ, Hoyle DC, Higgs PG (2001) RNA sequence evolution with secondary structure constraints: comparison of substitution rate models using maximum likelihood methods. Genetics 157:399–411
Schultz J, Maisel S, Gerlach D, Müller T, Wolf M (2005) A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11:361–364
Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M (2006) 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinf 7:498
Seibel PN, Müller T, Dandekar T, Wolf M (2008) Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res Notes 1:91
Sytsma KJ, Litt A, Zjhra ML, Pires JC, Nepokroeff M, Conti E, Walker J, Wilson PG (2004) Clades, clocks, and continents: historical and biogeographical analysis of Myrtaceae, Vochysiaceae, and relatives in the southern hemisphere. Int J Plant Sci 165(Suppl. 4):S85–S105
Thompson JD, Gibson T, Lewniak F, Jeanmougin F, Higgins D (1997) The ClustalX windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 24:4876–4882
Torres RA, Ganal M, Hemleben V (1990) GC balance in the internal transcribed spacers ITS1 and ITS2 of nuclear ribosomal DNA. J Mol Evol 30:170–181
Wilson PG, O′Brien MM, Heslewood MM, Quinn CJ (2005) Relationships within Myrtaceae sensu lato based on a matK phylogeny. Pl Syst Evol 251:3–19
Wuyts J, De Rijk P, Van de Peer Y, Winkelmans T, De Wachter R (2001) The European large subunit ribosomal RNA Database. Nucl Acids Res 29:175–177
Zucker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res 31:3406–3415
Acknowledgments
This research was funded by the Comisión Nacional de Investigación Científica y Tecnológica, Gobierno de Chile (CONYCIT, 24090098) and the University of Concepción (DIUC 209.111.054-1.0). The herbaria ASU, CONC, K, MO, RB, and the Botanical Garden of the University of Vienna provided material for DNA extraction. We thank Leslie Landrum for cooperation in sending samples for extraction, academic discussions, and critical reading of the manuscript. Eva Lucas and Edith Karpino (Jodrell Laboratory, Royal Botanic Garden, Kew), sent us DNA samples. División de Investigación Bogotá (DIB-Universidad Nacional de Colombia) and B. Matthew, C. Michael, and B. Erin from American Journal Expert for translations into English. We thank to anonymous reviewers for their suggestions and comments on the manuscript. JM received a Grant from the University of Concepción-Programa de Mejoramiento de la Calidad y la Equidad de la Educación Superior (MECESUP, UCO0708) and funding from CONICYT and Universidad Nacional de Colombia for a stay in the Department of Systematic and Evolutionary Botany, University of Vienna. TFS was partially financed by Austrian Science Fund (FWF) Grant No. P21723-B16.
Author information
Authors and Affiliations
Corresponding author
Appendix 1
Appendix 1
Taxon: collector and number (herbarium) (GenBank registration number for ITS, ETS)
Acca sellowiana (O. Berg) Burret: Lucas 205 (K) (AM234067, AM489888). Algrizea macrochlamys (DC.) Proença and Nic Lughadha: Giulietti and Harley 1648 (K) (AM234126, AM489890). Amomyrtus meli (Phil.) D. Legrand and Kausel: RBGE 1996-1083 (E) (AM234069, AM489891). Blepharocalyx cruckshankii (Hook. and Arn.) Nied.: Murillo 4219 (CONC) (JN660907, JN660857). Blepharocalyx salicifolius (Kunth) O. Berg: Landrum 11232 (ASU) (JN660936, JN660886). Blepharocalyx salicifolius (Kunth) O. Berg: Negritto 927 (CONC) (JN660935, JN660885). Calyptranthes concinna DC.: Lucas 74 (K) (AM234103, AM489898). Calyptranthes thomasiana O. Berg: Pollard 1195 (K) (AM234106, AM489901). Campomanesia pubescens O. Berg Brazil: Farinaccio and Costa 1669 (K) (AM234077, AM489903). Campomanesia guazumifolia (Cambess.) O. Berg Cult. RBG Sydney: Belsham M86; K (AM489902, AM234076). Decaspermum humile (G. Don) A.J. Scott: Belsham M82 (OTA) (AM234128, AM489905). Eugenia latifolia Aubl.: Prevost 4707 (K) (AM234091, AM489913). Eugenia stictosephala Kiaersk.: Zappi 406 (K) (AM234086, AM489908). Eugenia uniflora L.: Lucas 207 (K) (AM234088, AM489910). Gomidesia flagellaris (D. Legrand) Sobral: Lucas 83 (K) (AM234113, AM489918). Gomidesia tijucensis (Kiaersk.) D. Legrand: Zappi 305 (K) (AM234110, AM489915). Gossia inophloia (J.F. Bailey and C.T. White) Snow and Guymer: Belsham M79 (OTA) (AM234131, AM489919). Legrandia concinna (Phil.) Kausel: RBGE 1999-0656 (E) (AM234072, AM489921). Lophomyrtus obcordata (Raoul) Burret: Belsham M41 (OTA) (AM234146, AM489924). Luma apiculata (DC.) Burret: Murillo 4205 (CONC) (JN660910, JN660860). Luma chequen (Molina) A. Gray: Landrum 7873 (CONC) (JN660911, JN660861). Marlierea eugeniopsoides (Kausel) D. Legrand: Lucas 61 (K) (AM234107, AM489928). Marlierea obscura O. Berg: Lucas 88 (K) (AM234109, AM489930). Marlierea suaveolens Cambess.: Lucas 85 (K) (AM234108, AM489929). Metrosideros perforata (J.R. and G. Forst.) A. Rich.: Lucas 209 (K) (AM234141, AM489931). Myrceugenia alpigena (DC.) Landrum: Lucas 167 (K) (JN660892, JN660842). Myrceugenia alpigena (DC.) Landrum var. fuliginea (O. Berg) Landrum: G. Hatschbach 59697 (ASU) (JN660891, JN660841). Myrceugenia alpigena (DC.) Landrum var longifolia (Burret) Landrum: R. Harley 26218 (ASU) (JN660893, JN660843). Myrceugenia brevipedicellata (Burret) D. Legrand and Kausel: Landrum 2830 (ASU) (JN660894, JN660844). Myrceugenia campestris (DC.) D. Legrand and Kausel: R. Kummrow 2940 (ASU) (JN660895, JN660845). Myrceugenia chrysocarpa (O. Berg) Kausel: Landrum 8166 (CONC) (JN660896, JN660846). Myrceugenia colchaguensis (Phil.) Navas: Landrum 8033 (CONC) (JN660897, JN660847). Myrceugenia correifolia (Hook. and Arn.) O. Berg; Teillier 5360 (CONC) (JN660901, JN660851). Myrceugenia cucullata D. Legrand: Wasum 105 (ASU) (JN660898, JN660848). Myrceugenia euosma (O. Berg) D. Legrand: Soares 715 (ASU) (JN660899, JN660849). Myrceugenia exsucca (DC.) O. Berg: Murillo 4217 (CONC) (JN660900, JN660850). Myrceugenia fernandeziana (Hook. and Arn.) Johow: Stuessy 15283 (CONC) (JN660903, JN660853). Myrceugenia franciscensis (O. Berg) Landrum: Miyagi 357 (ASU) (JN660902, JN660852). Myrceugenia gertii Landrum: E. Barbosa 948 (ASU) (JN660904, JN660854). Myrceugenia glaucescens (Cambess.) D. Legrand and Kausel: Landrum 11231 (ASU) (JN660905, JN660855). Myrceugenia kleinii D. Legrand and Kausel: Cordeiro 734 (ASU) (JN660906, JN660856). Myrceugenia lanceolata (Juss. ex J. St.-Hil.) Kausel: Mihoc 6220 (CONC) (JN660908, JN660858). Myrceugenia leptospermoides (DC.) Kausel: Murillo 4214 (CONC) (JN660909, JN660859). Myrceugenia miersiana (Gardner) D. Legrand and Kausel: Lucas 164 (K) (JN660912, JN660862). Myrceugenia myrciodes (Cambess.) O. Berg var. acrophylla (O. Berg) D. Legrand: Ribas 229 (ASU) (JN660913, JN660863). Myrceugenia myrcioides (Cambess.) O. Berg: Lucas 503 (K) (JN660915, JN660865). Myrceugenia myrtoides O. Berg: Rossato 47 (MO) (JN660919, JN660869). Myrceugenia obtusa (DC.) O. Berg: Brownless 1227 (CONC) (JN660916, JN660866). Myrceugenia ovalifolia (O. Berg) Landrum: Lucas 259 (K) (JN660917, JN660867). Myrceugenia ovata (Hook. and Arn.) O. Berg var. acutata (D. Legrand) Landrum: Chagas 1979 (ASU) (JN660918, JN660868). Myrceugenia ovata (Hook. and Arn.) O. Berg var. nannophylla (Burret) Landrum: Mihoc 5162 (CONC) (JN660920, JN660870). Myrceugenia ovata (Hook. and Arn.) O. Berg var. ovata: Gardner 19 (CONC) (JN660922, JN660872). Myrceugenia ovata (Hook. and Arn.) O. Berg var. regnelliana (O. Berg) Landrum: Silva 18 (ASU) (JN660921, JN660871). Myrceugenia ovata (Hook. and Arn.) O. Berg var. regnelliana (O. Berg) Landrum: Souza 10621 (ASU) (-, JN660889). Myrceugenia oxysepala (Burret) D. Legrand and Kausel: Ribas 2234 (ASU) (JN660923, JN660873). Myrceugenia parvifolia (DC.) Kausel: Landrum 5916 (CONC) (JN660924, JN660874). Myrceugenia pilotantha (Kiaersk.) Landrum: Lucas 230 (K) (JN660925, JN660875). Myrceugenia pilotantha (Kiaersk.) Landrum: Lohmann 35 (ASU) (JN660926, JN660876). Myrceugenia pinifolia (F. Phil.) Kausel: Gardner 164 (CONC) (JN660927, JN660877). Myrceugenia planipes (Hook. and Arn.) O. Berg: Aedo 7378 (CONC) (JN660928, JN660878). Myrceugenia reitzii D. Legrand and Kausel: Barbosa 945 (ASU) (JN660937, JN660887). Myrceugenia rufa (Colla) Skottsb. ex Kausel: Teillier 150795 (CONC) (JN660929, JN660879). Myrceugenia rufescens (DC.) D. Legrand and Kausel: Lucas 469 (K) (JN660930, JN660880). Myrceugenia schulzei Johow: Ruiz 8266 (CONC) (JN660938, JN660888). Myrceugenia seriatoramosa (Kiaersk.) D. Legrand and Kausel: Silva 2358 (MO) (JN660932, JN660882). Myrceugenia smithii Landrum: García 533 (ASU) (JN660931, JN660881). Myrcia bicarinata (O. Berg) D. Legrand: Lucas 71 (K) (AM234121, AM489945). Myrcia coumeta (Aubl.) DC.: Lucas 107 (K) (AM234123, AM489947). Myrcia laricina (O. Berg) Burret ex Luetzelb.: Mello Silva 1713 (K) (AM234118, AM489942). Myrcia laruotteana Cambess.: Mello Silva 1705 (K) (AM234115, AM489939). Myrcia pubipetala Miq.: Lucas 86 (K) (AM234114, AM489938). Myrcia rostrata DC.: Lucas 73 (K) (AM234122, AM489946). Myrcia saxatilis (Amsh.) McVaugh: Lucas 98 (K) (AM234119, AM489943). Myrcia tomentosa (Aubl.) DC.: Soares 752 (K) (AM234116, AM489940). Myrcia venulosa DC.: Cruz and Cordeiro 195 (K) (AM234125, AM489949). Myrcianthes cisplatensis (Cambess.) O. Berg: Landrum 11233 (ASU) (JN660914, JN660864). Myrcianthes pseudomato (D. Legrand) McVaugh: Beck 9667 (K) (AM234100, AM489951). Myrciaria cauliflora O. Berg: Lucas 210 (K) (AM234093, AM489952). Myrteola nummularia (Lam.) O. Berg: RBGE 1996-1096 € (AM234068, AM489954). Myrtus communis L.: Botanical Garden Austria (JN660890, JN660840). Neomitranthes cordifolia (D. Legrand) D. Legrand: Forster 1011 (ESA) (AM489410, AM489413). Neomyrtus pedunculata (Hook. f.) Burret: Belsham M42 (OTA) (AM234144, AM489956). Pimenta pseudocaryophyllus (Gomes) Landrum: Lucas 161 (K) (AM234083, AM489960). Pimenta racemosa (Mill.) J.W. Moore: Holst 8866 (K) (AM234082, AM489959). Plinia pauciflora M.L. Kawas. and B. Holst: Mazine 957 (ESA) (AM489411, AM489414). Psidium cattleianum Sabine: Lucas 213 (K) (AM234080, AM489962). Psidium cinereum Mart. ex DC.: Da Silva and Farias, 4535 (K) (AM234079, AM489961). Psidium guajava L.: ASU199194 (ASU) (AY487283, AY454126). Rhodamnia rubescens (Benth.) Miq.: Belsham M83 (OTA) (AM234127, AM489963). Rhodomyrtus psidioides (G. Don) Benth.: Belsham M72 (OTA) (AM234134, AM489965). Siphoneugena guilfoyleana C. Proença: Lucas 70 (K) (AM234085, AM489966). Ugni molinae Turcz.: Murillo 4213 (CONC) (JN660933, JN660883). Ugni selkirkii (Hook. and Arn.) O. Berg: F. Gardner 31 (CONC) (JN660934, JN660884). Xanthomyrtus montivaga A.J. Scott: Lucas 16 (K) (AM234147, AM489971).
Rights and permissions
About this article
Cite this article
Murillo-A, J., Stuessy, T.F. & Ruiz, E. Phylogenetic relationships among Myrceugenia, Blepharocalyx, and Luma (Myrtaceae) based on paired-sites models and the secondary structures of ITS and ETS sequences. Plant Syst Evol 299, 713–729 (2013). https://doi.org/10.1007/s00606-012-0754-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-012-0754-x