Abstract
In this study, we evaluate the impact of fossil assignments and different models of calibration on divergence time estimates carried out as Bayesian analyses. Estimated ages from preceding studies and liverwort inclusions from Baltic amber are used as constraints on a molecular phylogeny of Cephaloziineae (Jungermanniopsida) obtained from sequences of two chloroplast coding regions: rbcL and psbA. In total, the comparison of 12 different analyses demonstrates that an increased reliability of the chronograms is linked to the number of fossils assigned and to the accuracy of their assignments. Inclusion of fossil constraints leads to older ages of most crown groups, but has no influence on lineage through time plots suggesting a nearly constant accumulation of diversity since the origin of Cephaloziineae in the early to Middle Jurassic. Our results provide a note of caution regarding the interpretation of chronograms derived from DNA sequence variation of extant species based on a single calibration point and/or low accuracy of the assignment of fossils to nodes in the phylogeny.
Similar content being viewed by others
Introduction
DNA-sequence-based divergence time estimates are now widely employed to study the evolution of various lineages of animals, fungi and plants. The application of these methods resulted in a dramatic improvement of our understanding of evolutionary events and processes, especially in lineages with a poor and patchy fossil record (Hedges and Kumar 2009). However, these results are often controversial, especially when comparisons with the fossil record can be drawn (e.g. Donoghue and Benton 2007; Kenrick et al. 2012; Parham et al. 2012). These controversies are partly caused by confusion about the accuracy of the information obtained by DNA sequence-based or fossil-based reconstruction of lineage histories and ages (Kenrick et al. 2012).
Arguably, one of the most important sources of inaccuracy in molecular-based divergence time estimates is the need to calibrate the molecular clocks using fossil evidence (Donoghue and Benton 2007; Hedges and Kumar 2009; Parham et al. 2012). Other approaches, such as estimates of geological events have been considered as alternatives to fossil-based calibrations, but their usage is restricted to certain cases and could result in misleading estimates (Renner 2005). Early methods of molecular clock dating used single calibration points despite the fact that the age of a fossil is nearly always documented as a time interval. Improvement has been achieved by the introduction of methods that employ minimum and maximum age constraints beside one or several calibration points (Ho 2007; Benton et al. 2009; Inoue et al. 2010; Magallon 2010; Heled and Drummond 2012; Lukoschek et al. 2012; Sauquet et al. 2012). The most recent innovations allow us to integrate the information given by fossil evidence in form of probability distributions. This kind of calibration is usually part of relaxed molecular clock methods that estimate divergence times based on the DNA sequence data using one or several genomic regions either as a single or several partitions. They allow the integration of several fossil constraints in a single analysis, but require to study the impact of fossil assignments on the sister lineages of the calibrated clades in the tree very careful (Benton et al. 2009; Inoue et al. 2010; Magallon 2010; Parham et al. 2012).
Most authors agree on the importance of the calibration and constraining of molecular clocks with the fossil record (Benton et al. 2009; Hedman 2010; Inoue et al. 2010; Magallon 2010; Pyron 2010; Wilkinson et al. 2011; Lukoschek et al. 2012; Sauquet et al. 2012) and various methods have been made available to integrate this information (Ho 2007; McCormack et al. 2010; Heled and Drummond 2012). So far, relatively few studies explored the impact of these parameters on the estimates obtained from these analyses (e.g., Jacques et al. 2007; Sauquet et al. 2012). However, such studies are required to educate us about the limitations of the currently available methods and to develop guidelines of the employment of parameter models and fossil information (Near and Sanderson 2004; Hedman 2010; Inoue et al. 2010; Lukoschek et al. 2012).
Here, we use a main clade of leafy liverworts (Jungermanniidae), the Cephaloziineae, as a model to study the impact of fossil constraints. Generally, liverworts have a rather limited fossil record (Krassilov and Schuster 1984; Taylor et al. 2009), but they are often found as amber inclusions with a very high quality in character preservation (Grolle and Meister 2004a). Unfortunately, amber occurs discontinuously in space and time (Grimaldi 1996). This preservation bias in form of gaps in the record over time has to be taken into consideration when using amber fossils to infer the diversification of organisms (Smith 2001; Pyron 2010; Dornberg et al. 2011; Lloyd et al. 2012). DNA sequence-based divergence time estimates are potentially capable to overcome this shortcoming by integrating the discontinuous amber fossil record into the framework provided by either an already dated phylogeny or preferably into simultaneously inferred divergence time estimates.
The high quality of the preservation of amber fossils allows for a description of many morphological characters that are phylogenetically or taxonomically informative. The exquisite preservation enables the application of roles to the assignment proposed as best practice (Parham et al. 2012) despite some uncertainties concerning the taxonomic interpretation and age of the fossils. Several studies focused on the relationships of amber inclusions of liverworts to extant taxa (Grolle 1983, 1985, 1993; Gradstein 1993; Grolle and Schmidt 2001; Grolle and Meister 2004a, b; Heinrichs and Schmidt 2010; Heinrichs et al. 2011, 2012b). Thus, these fossils can be potentially assigned to particular nodes in the phylogeny of the studied lineage of liverworts and some studies reconstructed the diversification of selected liverwort lineages using either a DNA clock only approach or a DNA plus amber fossil approach (Hartmann et al. 2006, 2007, 2009b; Wilson et al. 2007; Devos and Vanderpoorten 2009). Amber inclusions are, however, not the only source of liverwort fossils (Krassilov and Schuster 1984). Some liverworts fossils have been recorded with different taphonomic background such as Sinolejeunea from the Middle Jurassic Yima Formation (Yang and Wu 2010) or anatomical preserved remnants of leafy liverworts from the Eocene of Canada (Steenbock et al. 2011).
Molecular phylogenetic studies indicate that the liverworts (Marchantiophyta) have separated from other lineages in the earliest diversification of plants on land (Qiu et al. 2006). Heinrichs et al. (2007) estimated the age of the leafy liverworts (Jungermanniidae) to the late Carboniferous (308.7 ± 7.8 Ma), but slightly older ages may need to be considered given the uncertainty of divergence time estimates of the oldest splits of liverworts (Clarke et al. 2011; Kenrick et al. 2012).
In this study, we explore the integration of the amber fossil record in studies focusing on the diversification of a derived lineage of liverworts, the Jungermannialean lineage Cephaloziineae (Crandall-Stotler et al. 2009), which comprises the families Cephaloziaceae, Cephaloziellaceae, Hygrobiellaceae, Adelanthaceae s. l. and Scapaniaceae s. l. We are particularly interested in the impact of alternative assignments that reflect uncertainties in the interpretation of the fossils’ relationships (see also Sauquet et al. 2012). This study will also provide us with empirical evidence about the performance of recently established methods to estimate divergence times in the context of phylogenetic uncertainty concerning the relationships of the taxa preserved in the fossil record (Drummond et al. 2006). All analyses were carried out using BEAST, a Bayesian MCMC method capable to obtain time-measured phylogenies with confidence intervals using relaxed molecular clocks and various parameters to integrate information about the evolutionary processes and the fossil record (Drummond et al. 2006). The main focus of this study is on the performance of relaxed clock methods using state-of-the-art calibration tools (Ho 2007; Battistuzzi et al. 2010; Sauquet et al. 2012). In particular, we investigate the impact of different calibrations on the estimates of diversification rate changes through time.
Materials and methods
Taxon sampling and assembling of DNA sequence data
For this study, we obtained chloroplast DNA rbcL- and psbA-sequences of most genera that are assigned to the Cephaloziineae based on the morphological studies or on the molecular phylogenetic results of several preceding works (Forrest et al. 2006; He-Nygrén et al. 2006; de Roo et al. 2007; Heinrichs et al. 2007; Feldberg et al. 2009, 2010; Vilnet et al. 2010). In addition to a DNA dataset of Adelanthaceae s. l. (Feldberg et al. 2010), we obtained rbcL and psbA sequences of related lineages via DNA sequencing or GenBank.
For DNA sequencing plant tissue from the distal portions of a few shoots was isolated from herbarium collections. Total genomic DNA was purified using Invisorb Spin Plant Mini Kit (Invitek, Berlin, Germany) prior to amplification. Protocols for PCR were carried out as described in the previous publications: rbcL from Hentschel et al. (2006), and psbA from Forrest and Crandall-Stotler (2004).
Bidirectional sequences were generated using an MegaBACE 1000 automated sequencing machine using DYEnamic ET Primer DNA Sequencing Reagent (Amersham Biosciences, Little Chalfont, UK). Newly generated sequences were assembled and edited using SeqAssem (Hepperle 2004).
Initial phylogenetic analyses
All sequences (Table 1) were aligned manually in BioEdit version 5.0.9 (Hall 1999). Ambiguous positions were excluded from the alignment; missing nucleotides in the aligned sequences were coded as missing. jModelTest 0.1 (Posada 2008) was employed to choose nucleotide substitution models each for the rbcL as well as the psbA dataset and the combined matrices. The Akaike information criterion supported the GTR + Γ + I model as the best fit for both partitions and also for the unpartitioned dataset. Topological congruence of the two datasets was explored by comparing visually phylogenetic trees obtained from maximum likelihood-based bootstrap analyses with Garli 0.96 (Zwickl 2006).
All divergence time analyses were carried out using BEAST package version 1.5.4 (Drummond and Rambaut 2007). With BEAUti 1.5.4 (BEAST package) two partitions were created, one for each marker, in addition to a combined dataset. Because previous analyses rejected a strict clock for the leafy liverworts (Heinrichs et al. 2007), we used the uncorrelated relaxed clock model (Drummond et al. 2006) with a lognormal distribution of rates estimated during the analyses. For each analysis, one run with 100 million generations, and sampling of every 10,000th generation was undertaken. All analyses were carried out with the models selected by jModelTest 0.1 (Posada 2008), with the age estimates and a birth–death model implemented. Tracer 1.5 (Rambaut and Drummond 2003–2009) was used to check the effective sampling size (ESS) for all parameters. The results were considered reliable when the ESS exceeded 500 for all parameters. Age estimates were summarized in a mean consensus chronogram built with TreeAnnotator 1.5.4 (BEAST package) after exclusion of trees recovered during the burn-in-phase. FigTree 1.3.1 (Rambaut 2006–2009) was used to display the consensus tree created via TreeAnnotator. All analyses were carried out with calibrations described below. To achieve comparability, only calibrations were modified.
Assignment of fossil evidence
We tested several approaches to calibrate the tree. First, a series of analyses with only an age constraint for the root node was employed. BEAST performs more reliable under the assumption of a basal constraint (Drummond et al. 2006; McCormack et al. 2010), but since no fossil is known that could serve as maximum age constraint the age for this node was taken from a previous divergence time estimate for liverworts (Heinrichs et al. 2007). We chose the minimum age of 158.0–500.0 Ma for the analyses with uniform distributed priors. Based on the results of this analysis, we chose the estimated age of 171.1 ± 8.3 Ma and a normal distributed prior for following analyses.
Beside analyses with single root node calibrations, we carried out several analyses with additional fossil-based calibrations. Three inclusions from Eocene Baltic amber (35–48 Ma; Standke 1998) are applicable to the tree: Scapania hoffeinsiana Grolle, Lophozia kutscheri Grolle, and Cylindrocolea dimorpha (Casp.) Grolle (Grolle and Schmidt 2001; Grolle and Meister 2004a, b; Frahm 2006). All three fossils are sufficiently well preserved and their taxonomic relationships have been thoroughly discussed by Grolle and Meister (2004a).
Both gametophyte and sporophyte of S. hoffeinsiana have been well preserved and indicated it as a member of the crown-group of Scapania. It resembles Scapania (subg. Scapania) umbrosa (Schrad.) Dumort. in several aspects (Grolle and Schmidt 2001; Grolle and Meister 2004a). The perianth is dorsally eplicate, a common character of members of the derived subgenus Scapania. Scapania developed from an ancestor with a plicate perianth, however, not only most members of the derived subg. Scapania have eplicate perianths, but also some early diverging species (Heinrichs et al. 2012a). We assume that the different perianth-forms of Scapania derived from the plicate perianth type and that the eplicate perianth is an apomorphy of the crown group. In combination with the leaf characters of S. hoffeinsiana it indicates a position within S. subg. Scapania and can be distinguished from independently evolved similar structures in some earlier diverging lineages of Scapania. Therefore, S. hoffeinsiana may not only be assigned as the ancestor of the whole genus Scapania, but also as the ancestor of the subgenus Scapania. We performed several analyses with only the basal constraint and different assignments of S. hoffeinsiana to test if this significantly influenced the ages of the sister lineages.
For the other fossils, it was not possible to test varying assignments to higher or lower ranking clades, so we placed them near to the most similar extant relative to perform analyses with all three fossils. Based on the morphological treatment in Grolle and Meister (2004a) C. dimorpha was assigned to crown group node B containing C. recurvifolia (Steph.) Inoue, and L. kutscheri was assigned to the base of the Barbilophozia clade, since it is most similar to Barbilophozia hatcheri (A. Evans) Loeske.
Divergence times estimates and lineages through time plots
Three of the 12 analyses were performed with a root node-constraint only: 1.A: a partitioned dataset with uniform prior distribution of 158.0–500.0 Ma, 2.A: a partitioned dataset with normal distributed prior distribution of 171.1 ± 8.3 Ma and 3.A: an un-partitioned dataset with normal prior distribution of 171.1 ± 8.3 Ma.
The Eocene amber fossils were added as uniform minimum age constraints of 35.0–100.0 Ma. Lineages through time plots for the ingroup (A) of the analyses 2.A and 2.D were performed with Tracer 1.5.
Results
Divergence time estimates using a single age constraint with different prior-models for the Cephaloziineae
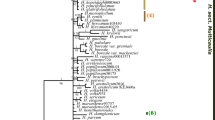
The three analyses (1.A, 2.A, and 3.A; Tables 2, 3, 4) provide different age estimates for Cephaloziineae (node A*; Figs. 1, 2). The partitioned dataset with a uniform prior recovers the oldest estimate with a node age of 208.68 Ma and a confidence interval of 158.01–371.77 Ma (Table 2; Fig. 3). The age of node A fluctuates strongly in different runs with the same settings (trees not shown) and the confidence intervals are quite broad (Fig. 3).
The analyses with normal constraints produce younger ages and different runs do not show the fluctuation seen in analysis 1.A (Tables 2, 3, 4; Fig. 3). In analysis 2.A node A* is estimated to 171.84 Ma (confidence interval 153.03–185.76 Ma) and in analysis 3.A it is estimated to 180.01 Ma (confidence interval 153.88–185.96 Ma). Unpartitioned and partitioned analyses with a normal distributed basal calibration show rather narrow confidence intervals, in contrast to the results in analysis 1.A.
Clades with assignable amber inclusions (calibration nodes B*, C*, D1* and D2*; Figs. 1, 2) are estimated younger than the fossils indicate. The mean age of the node B* within the Cephaloziellaceae ranges between 25.17 Ma in analysis 1.A, 14.31 Ma in analysis 2.A and 22.66 Ma in analysis 3.A (Tables 2, 3, 4). The mean age of node C* (crown group Barbilophozia) ranges between 30.42 Ma (analysis 1.A), 17.22 Ma (analysis 2.A) and 14.11 Ma (analysis 3.A). Node D1* (crown group Scapania) shows a similar pattern; the mean ages are 24.39 Ma (analysis 1.A), 21.09 Ma (analysis 2.A) and 28.25 Ma (analysis 3.A), while the age of node D2* (crown group S. subg. Scapania) ranges between 20.92 Ma (analysis 1.A), 18.7 Ma (analysis 2.A) and 22.71 Ma (analysis 3.A).
Divergence time estimates using different models of root node constraints and different fossil calibrations
Additional uniform minimum age constraints have a strong impact on the divergence time estimates (analyses 1–3.B) with a constraint for node D2* (crown group Scapania subg. Scapania); analyses 1–3.C with a constraint for node D1* (crown group Scapania); analyses 1–3.D with constraints for nodes D2*, B* [Cylindrocolea plus Cephaloziopsis intertexta (Gottsche) R.M.Schust. and Cephalozia microphylla Steph.] and C* (crown group Barbilophozia). The results are summarized in Tables 2, 3, 4.
The analysis 1.B yields older node ages than analysis 1.C. If the age constraint is placed in Scapania subg. Scapania the genus becomes older than if the constraint is placed in Scapania s. l. (76.08 vs. 36.46 Ma; Table 2). This difference does not only affect Scapania, but also other nodes. The Cephaloziaceae (C.0) have an estimated age of 396.08 Ma in 1.B and of 151.58 Ma in 1.C, the Adelanthaceae (AD.0) have an estimated age of 235.2 Ma in 1.B and 106.69 Ma in 1.C, the Cephaloziellaceae (CE.0) have an estimated age of 95.23 Ma in 1.B and 45.11 Ma in 1.C, and the Scapaniaceae s. l. (S.0) have an estimated age of 156.22 Ma in 1.B and 94.5 Ma in 1.C (Fig. 3; see Table 2 for the ages of other groups). The oldest node ages are estimated in analysis 1.D with all fossil constraints applied. Here, the Cephaloziaceae (C.0) have an age of 349.05 Ma, the Adelanthaceae (AD.0) of 209.89 Ma, the Cephaloziellaceae (CE.0) of 90.31 Ma, and the Scapaniaceae s. l. (S.0) of 174.53 Ma (Fig. 3; see Table 2 for the ages of other groups).
In analyses 2.B/3.B Cephaloziaceae (C.0) have an age of 165.38/157.08 Ma and in analyses 2.C/3.C of 158.87/165.16 Ma. Adelanthaceae (AD.0) are estimated to 109.18/97.07 Ma in analyses 2.B/3.B and 106.72/82.42 Ma in analyses 2.C/3.C; Cephaloziellaceae (CE.0) are estimated to 51.46/48.14 Ma in analyses 2.B/3.B and 56.02/48.24 Ma in analyses 2.C/3.C. The Scapaniaceae s. l. crown group (S.0) has an age of 94.73/89.78 Ma in analyses 2.B/3.B and 87.68/94.29 Ma in analyses 2.C/3.C (Fig. 3; see Tables 3, 4 for the ages of other nodes). Node ages in analyses 2.B/3.B and 2.C/3.C are more similar than in analyses 1.B/1.C. The nodes B*, C*, D1* and D2* are often estimated younger than the fossils indicate.
In analyses 2.D (Fig. 2)/3.D with all fossil constraints applied the Cephaloziaceae (C.0) have an estimated age of 147.71/158.59 Ma, the Adelanthaceae (AD.0) have an age of 142.92/116.81 Ma, the Cephaloziellaceae (CE.0) of 69.89/66.79 Ma, and the Scapaniaceae s. l. (S.0) of 108.11/100.76 Ma (Fig. 3; see Table 3, 4 for the ages of other groups).
Partitioned versus non-partitioned analyses and lineages through time plots
There are no significant differences between the partitioned and the unpartitioned analyses (Fig. 3; Tables 2, 3, 4). Lineage through time plots (Figs. 1, 2) of all analyses showed evidence for nearly constant diversification rates.
Discussion
Uncertainty of assignment of fossils
The assignment of fossil taxa to nodes reconstructed using extant taxa proves to be a challenge of assumptions. The division of stem versus crown group assignments is among the most widely accepted but still poorly resolved issues (Rutschmann et al. 2007; Gernandt et al. 2008). The distinction is logically consistent but both estimates may create substantially different time frames in which macro-evolutionary hypotheses such as the Gondwanan vicariance hypothesis (Raven and Axelrod 1974) are inferred. A far less addressed issue is the interpretation of the morphological characters used to assign the fossil to a particular node in the tree. Ideally, the character used represents an apomorphy, such as the lindsaeoid root anatomy recovered in Cretaceous fern fossils (Schneider and Kenrick 2001; Schneider et al. 2004). However, many fossils do not show characters that represent unambiguous apomorphic character states. The vast majority of characters preserved in fossils show a strong pattern of homoplasy as a result of factors such as exhaustion of morphological variation, frequent parallelism and fossil bias in the preservation of particular structures (e.g., Wagner 2000; Schneider 2007; Schneider et al. 2009). Thus, assignments are often based on the combination of characters of which each one separately is ambiguous.
Furthermore, assignments are often done without careful consideration of the evolution of characters preserved in the fossil under study. The use of plotting morphological characters onto phylogenetic trees based on the extant taxa may be a realistic approach to overcome these questions (Schneider et al. 2009). However, also this approach has its own limits because it assumes that the reconstruction of the character evolution using extant taxa alone may be congruent to the true evolution of this character through time including both extant and extinct taxa (e.g. Schneider 2007).
The fossil liverworts used in this study are excellent examples to illustrate this problem. There are many homoplasies in the relatively uniform vegetative morphology of liverworts (Crandall-Stotler et al. 2005; Hentschel et al. 2006; Heinrichs et al. 2007) and also the reproductive structures, which are generally better suited for evolutionary reconstruction, show sometimes signs of independent parallel evolution (Hentschel et al. 2006).
Furthermore, there are only few fossil liverworts with well-preserved female structures (Heinrichs et al. 2012c). S. hoffeinsiana is one of these rare amber fossils with perianth and sporophyte and, like mentioned above; it is not easy to assign it to an extant group. Like other members of Cephaloziineae the genus Scapania possesses a true perianth made up of adnate modified leaves. It is plicate or eplicate with both character states seen in early diverging Scapania lineages and the derived subg. Scapania (Heinrichs et al. 2012a). To cope with the tentativeness of the S. hoffeinsiana assignment we tested several possibilities, with great impact on the age of not only the genus Scapania, but also its sister groups (Figs. 1, 2, 3; Tables 2, 3, 4). Other fossil assignments are hampered by incongruence of morphology-based genus concepts and molecular phylogenies, as demonstrated by the paraphyly of the genus Cephaloziella with Cylindrocolea, Cephaloziopsis and Kymatocalyx nested in it (Figs. 1, 2). Consequently, deeper insights into the morphological evolution of the Cephaloziella lineage and the taxonomical value of certain character states require more comprehensive phylogenies with a dense sampling of species. These phylogenies will possibly allow for more reliable assignments of the related fossil Cylindrocolea/Cephaloziella dimorpha than currently possible.
Evaluating the impact of multiple constraints and character-rich models
Our results demonstrate the potential strong impact of fossil constraints, but also highlight the need to carefully assess the assignment of the fossil to prevent misleading results. We suggest a reciprocal illumination approach being the ideal method addressing this question by comparing carefully the age estimates obtained by analyses without and with the fossil constraint under question. For example, the omission of C. dimorpha and L. kutscheri resulted in a young age not only for the clades in question, but also for the rest of the tree.
Several clades were estimated much younger without the inclusion of the information from their fossil relatives. This result may indicate either problems with the interpretation of their taxonomic relationships or more likely limitations of the Bayesian methodology to estimate the age of these nodes.
Interestingly, the lineages through time plots of all analyses provide evidence for persistent diversification rates. The slower accumulation of species diversity towards the present is likely the result of the taxonomic sampling and therefore considered as an artefact. Thus, the reported issues concerning the absolute age estimate of some nodes had little impact on the reconstructed processes. However, this may be partly due to the lack of sensitivity of lineage through time plots. Other methods to infer diversification rates (e.g., Rabovsky 2010; Cusimano et al. 2012) may behave differently but this needs to be explored in future studies.
Towards the reconstruction of the evolutionary history of leafy liverworts
Despite the main objective of this study concerned the assessment of divergence times, the results provide also some new insights into the history of leafy liverworts. Previous studies on the divergence times of liverworts addressed the age of the main lineages (e.g., Heinrichs et al. 2007; Cooper et al. 2012), biogeographic hypotheses such as pseudo-Gondwana distributions (Hartmann et al. 2006; Heinrichs et al. 2009b), and the pattern of divergences of liverworts. The latter studies reported evidence for the conservation of diversification rates in liverworts for the most of the Mesozoic and the Cenozoic. Nearly constant diversification rates of liverworts were found by both extensive analyses of a species-based sampling of the derived leafy liverwort family Lejeuneaceae (Wilson et al. 2007) and a family-based sampling of liverworts (Fiz-Palacios et al. 2011). These reports are remarkable given the trend of strong fluctuations of species richness through time recovered in studies on the fossil record of vascular plants (Niklas et al. 1983; Lidgard and Crane 1988; Benton 1995; Lupia et al. 1999) and in divergence time analyses in angiosperms, gymnosperms, ferns, and mosses (Schneider et al. 2004, 2010a, b; Janssen et al. 2008; Schuettpelz and Pryer 2009; Shaw et al. 2010; Crisp and Cook 2011; Eiserhardt et al. 2011; Nagalingum et al. 2011; Smith et al. 2011). The Cenozoic is characterized not only by strong modifications of the continental surfaces such as the rise of the Himalaya (Wang et al. 2012) but also by significant climatic events such as the Eocene climate optimum, the late Eocene and early Oligocene cooling and the Miocene warming. None of these events appears to have significantly influenced the diversification rates of the predominantly tropical Lejeuneaceae (Wilson et al. 2007).
With the exception of the mainly tropical Adelanthaceae, the Cephaloziineae are more abundant in temperate and subarctic regions than in the inner tropics. The predominant occurrence of Cephaloziineae in temperate regions possessing more unstable climates strongly indicated this family as a candidate of a leafy liverwort lineage with fluctuations in diversification rates in response to climatic fluctuations. Nevertheless, the recovered pattern for Cephaloziineae is consistent with the hypothesis of rather consistent diversification rates in liverworts, as already demonstrated for the Lejeuneaceae family. However, the reported evidence may be inaccurate because the present study includes less than 20 % of the extant species diversity of Cephaloziineae. The limited sample may led to an oversampling of deep nodes and thus reduce the sensitivity of the analyses to detect changes in the diversification rates. The same holds true for the Lejeuneaceae study (Wilson et al. 2007). Thus, the current hypothesis of constant diversification rates within the leafy liverworts may be a result of sampling bias (Cusimano et al. 2012). Follow-up studies will thus need to address issues concerning the impact of taxon sampling and phylogenetic robustness, and should aim at a more comprehensive taxon sampling. These studies need to consider the limitations of analyses based solely on the extant species in the recovery of changes in speciation and extinction rates (Marshall 2008; Rabovsky 2010). The bias in studies using exclusively extant taxa may result in revisionist estimates of the evolutionary history of these lineages (Tarver and Donoghue 2011). Hence, studies are required testing the molecular dating-based hypothesis using the fossil record.
State-of-the-art and perspectives
Reconstructing the divergence times of liverworts is hampered by our limited knowledge on their early evolutionary history (Clarke et al. 2011; Kenrick et al. 2012), their sparse fossil record (Taylor et al. 2009) and uncertainties with regard to the age of many fossils (Iturralde-Vincent and MacPhee 1996). Our results are consistent with the previous studies reporting evidence that the usage of multiple-constraints requires careful consideration because of the difficulties in predicting the impact of each individual constrain in Bayesian analyses (Inoue et al. 2010). However, more empirical studies are required to elucidate the robustness versus sensitivity of these methods as required to develop clear guidelines for their usage.
The deviations of divergence times in our different analyses provide a note of caution regarding interpretations of chronograms produced for lineages without or with a sparse fossil record. The present state-of-the art already allows for balancing different macroevolutionary scenarios, such as vicariance or dispersal, but explicit correlations of diversification events of bryophytes with climatic fluctuations or orogeneses are still premature and deserve more reliable chronograms. Advancements in our knowledge of the fossil record are highly desirable to arrive at more reliable conclusions on their historical biogeography (Heinrichs et al. 2009a).
In recent years, the number of well-studied liverwort fossils has remarkably increased, especially for the Cenozoic. Integrating this enhanced fossil record in studies on the diversification of liverworts through time may enable us to overcome the limitations of extant taxa only approaches, especially the question of overlooked species turnovers and lineage replacements. The suggested integration of amber fossils into a phylogenetic framework may also enable us to overcome the core problem of the liverwort fossil record, the discontinuous occurrence of amber deposits in the Phanerozoic. Because the majority of liverwort fossils represents amber inclusions, the fossil record of this group is biased in several aspects: (1) fragmented in time periods with or without known amber deposits, (2) preservation of taxa occurring in habitats dominated by seed plants producing resins that are finally deposited as amber, and (3) climate conditions favouring the formation of forests producing huge amounts of resin exudates.
Improving analytical methods, increasing taxon sampling, and integration of a more comprehensive fossil record will likely lead to more reliable chronograms and allow for scrutinizing current hypotheses on a constant diversification of liverworts through time. These studies will allow to relate the liverwort diversification to the diversification of angiosperms, and will provide evidence for or against changes of liverwort lineages during the numerous rearrangements of terrestrial ecosystems in the late Mesozoic and early Cenozoic.
References
Battistuzzi FU, Filipski A, Hedges SB, Kumar S (2010) Performance of relaxed-clock methods in estimating evolutionary divergence times and their credibility intervals. Molec Biol Evol 27:1289–1300
Benton MJ (1995) Diversification and extinction in the history of life. Science 268:52–58
Benton MJ, Donoghue PCJ, Asher RJ (2009) Calibrating and constraining molecular clocks. In: Hedges SB, Kumar S (eds) Timetree of Life. University Press, Oxford, pp 35–86
Clarke JT, Warnock RCM, Donoghue PCJ (2011) Establishing a time-scale for plant evolution. New Phytol 192:266–301
Cooper EE, Henwood MJ, Brown EA (2012) Are the liverworts really that old? Cretaceous origins and Cenozoic diversifications in Lepidoziaceae reflect a recurrent theme in liverwort evolution. Biol J Linn Soc 107:425–441
Crandall-Stotler BJ, Forrest LL, Stotler RE (2005) Evolutionary trends in the simple thalloid liverworts. Taxon 54:299–316
Crandall-Stotler B, Stotler RE, Long DG (2009) Phylogeny and classification of the Marchantiophyta. Edinburgh J Bot 66:155–198
Crisp MD, Cook LG (2011) Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytol 192:997–1009
Cusimano N, Stadler T, Renner SS (2012) A new method for handling missing species in diversification analysis applicable to randomly or non-randomly sampled phylogenies. Syst Biol 61:785–792
de Roo RT, Hedderson TA, Söderström L (2007) Molecular insights into the phylogeny of the leafy liverwort family Lophoziaceae Cavers. Taxon 56:301–314
Devos N, Vanderpoorten A (2009) Range disjunctions, speciation, and morphological transformation rates in the liverwort genus Leptoscyphus. Evolution 63:779–791
Donoghue PCJ, Benton MJ (2007) Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol Evol 22:424–431
Dornberg A, Beaulie JM, Oliver JC, Near TJ (2011) Integrating fossil preservation biases in the selection of calibrations for molecular divergence time estimation. Syst Biol 60:519–527
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214 (http://beast.bio.ed.ac.uk)
Drummond AJ, Ho SY, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88
Eiserhardt WL, Rohwer JG, Russell SJ, Yesliyurt JC, Schneider H (2011) Evidence for radiations of cheilanthoid ferns in the Greater Cape Floristic region. Taxon 60:1269–1283
Feldberg K, Hentschel J, Bombosch A, Long DG, Váňa J, Heinrichs J (2009) Transfer of Gottschelia grollei, G. patoniae and Scaphophyllum speciosum to Solenostoma based on chloroplast DNA rbcL sequences. Pl Syst Evol 280:243–250
Feldberg K, Váňa J, Long DG, Shaw AJ, Hentschel J, Heinrichs J (2010) A phylogeny of Adelanthaceae (Jungermanniales, Marchantiophyta) based on nuclear and chloroplast DNA markers, with comments on classification, cryptic speciation and biogeography. Molec Phylogen Evol 55:293–304
Fiz-Palacios O, Schneider H, Heinrichs J, Savolainen V (2011) Diversification of land plants: insights from a family-level phylogenetic analysis. BMC Evol Biol 11:341
Forrest LL, Crandall-Stotler BJ (2004) A phylogeny of the simple thalloid liverworts (Jungermanniopsida, Metzgeriidae) as inferred from five chloroplast genes. Monogr Syst Bot Missouri Bot Gard 98:119–140
Forrest LL, Davis EC, Long DG, Crandall-Stotler BJ, Clark A, Hollingsworth ML (2006) Unraveling the evolutionary history of the liverworts (Marchantiophyta): multiple taxa, genomes and analyses. Bryologist 109:303–334
Frahm J-P (2006) Neue Moosfunde aus Baltischem Bernstein. Limprichtia 29:119–129
Gernandt DS, Magallon S, López GG, Flores OZ, Willyard A, Liston A (2008) Use of simultaneous analysis to guide fossil-based callibrations of Pinaceae phylogeny. Int J Pl Sci 169:1086–1099
Gradstein SR (1993) New fossil Hepaticae preserved in amber of the Dominican Republic. Nova Hedwigia 57:353–374
Grimaldi DA (1996) Amber. Window to the past. Abrams, New York
Grolle R (1983) Leucolejeunea antiqua n. sp., das erste Lebermoos aus Dominikanischem Bernstein. Stuttgarter Beitr Naturk, B 96:1–9
Grolle R (1985) Monograph of Frullania in Baltic amber. Prace Muz. Ziemi (Warsaw) 37:87–100
Grolle R (1993) Bryopteris bispinosa spec. nov. (Lejeuneaceae), ein weiteres Lebermoos in dominikanischem Bernstein. J Hattori Bot Lab 74:71–76
Grolle R, Meister K (2004a) The liverworts in Baltic and Bitterfeld amber. Weissdorn, Jena
Grolle R, Meister K (2004b) Lophozia kutscheri, a new hepatic (Jungermanniales) in Bitterfeld amber from central Germany. Bryologist 107:79–81
Grolle R, Schmidt A (2001) A fossil Scapania (Hepaticae) with perianth and capsule in Bitterfeld amber (Eocene) from Germany. Bryologist 104:362–366
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hartmann FA, Wilson R, Gradstein SR, Schneider H, Heinrichs J (2006) Testing hypotheses on species delimitations and disjunctions in the liverwort Bryopteris (Jungermanniopsida: Lejeuneaceae). Int J Pl Sci 167:1205–1214
Hedges SB, Kumar S (2009) Discovering the Timetree of Life. In: Hedges SB, Kumar S (eds) Timetree of Life. University Press, Oxford, pp 3–18
Hedman MH (2010) Constraints on clade ages from fossil outgroups. Paleobiol 36:16–31
Heinrichs J, Schmidt AR (2010) An inclusion of Frullania subgen. Diastaloba s.l. (Frullaniaceae, Porellales) in Dominican amber. Trop Bryol 31:142–156
Heinrichs J, Hentschel J, Wilson R, Feldberg K, Schneider H (2007) Evolution of leafy liverworts (Jungermanniidae, Marchantiophyta): estimating divergence times from chloroplast DNA sequences using penalized likelihood with integrated fossil evidence. Taxon 56:31–44
Heinrichs J, Hentschel J, Feldberg K, Bombosch A, Schneider H (2009a) Phylogenetic biogeography and taxonomy of disjunctly distributed bryophytes. J Syst Evol 47:497–508
Heinrichs J, Klugmann F, Hentschel J, Schneider H (2009b) DNA taxonomy, cryptic speciation and diversification of the Neotropical-African liverwort, Marchesinia brachiata (Lejeuneaceae, Porellales). Molec Phylogen Evol 53:113–121
Heinrichs J, Reiner-Drehwald ME, Feldberg K, Grimaldi DA, Nascimbene PC, von Konrat M, Schmidt AR (2011) Kaolakia borealis nov. gen. et sp. (Porellales, Jungermanniopsida): a leafy liverwort from the Cretaceous of Alaska. Rev Palaeobot Palynol 165:235–240
Heinrichs J, Bombosch A, Feldberg K, Kreier HP, Hentschel J, Eckstein J, Long D, Zhu R-L, Schäfer-Verwimp A, Schmidt AR, Shaw B, Shaw AJ, Váňa J (2012a) A phylogeny of the northern temperate leafy liverwort genus Scapania (Scapaniaceae, Jungermanniales). Molec Phylogen Evol 62:973–985
Heinrichs J, von Konrat M, Grabenhorst H, Schmidt AR (2012b) The sporophyte of the Paleogene liverwort Frullania varians Caspary. Fossil Rec 15:115–120
Heinrichs J, Reiner-Drehwald ME, Feldberg K, von Konrat M, Hentschel J, Váňa J, Grimaldi DA, Nascimbene PC, Schmidt AR (2012c) The leafy liverwort Frullania (Jungermanniopsida) in the Cretaceous amber forest of Myanmar. Rev Palaeobot Palynol 169:21–28
Heled J, Drummond AJ (2012) Calibrated tree priors for relaxed phylogenetics and divergence time estimation. Syst Biol 61:138–149
Hentschel J, Wilson R, Burghardt M, Zündorf H-J, Schneider H, Heinrichs J (2006) Reinstatement of Lophocoleaceae (Jungermanniopsida) based on chloroplast gene rbcL data: exploring the importance of female involucres for the systematics of Jungermanniales. Pl Syst Evol 258:211–226
He-Nygrén X, Juslén A, Ahonen I, Glenny D, Piippo S (2006) Illuminating the evolutionary history of liverworts (Marchantiophyta)—towards a natural classification. Cladistics 22:1–31
Hepperle D (2004) SeqAssem©. A sequence analysis tool, contig assembler and trace data visualization tool for molecular sequences. Win32-Version. Distributed by the author via: http://www.sequentix.de
Ho SYW (2007) Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biol 38:409–414
Inoue J, Donoghue PCJ, Yang Z (2010) The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst Biol 59:74–89
Iturralde-Vincent MA, MacPhee RDE (1996) Age and paleogeographical origin of Dominican amber. Science 273:1850–1852
Jacques FMB, Gallut C, Vignes-Lebbe R, Bagils RZI (2007) Resolving phylogenetic reconstruction in Menispermaceae (Ranunculales) using fossils and a novel statistical test. Taxon 56:379–392
Janssen T, Bystriakova N, Rakotondrainibe F, Coomes D, Labat J-N, Schneider H (2008) Neoendemism in Madagascan scaly tree ferns results from recent, coincident diversification bursts. Evolution 62:1876–1889
Kenrick P, Wellman CH, Schneider H, Edgecombe GD (2012) A timeline for terrestrialisation—consequences for the carbon cycle in the Palaeozoic. Philos Trans, Ser B 376:519–536
Krassilov VA, Schuster RM (1984) Paleozoic and Mesozoic fossils. In: Schuster RM (ed) New manual of bryology. The Hattori Botanical Laboratory, Nichinan, pp 1172–1193
Lidgard S, Crane PR (1988) Quantitative analyses of the early angiosperm radiation. Nature 331:344–346
Lloyd GT, Young JR, Smith AB (2012) Taxonomic structure of the fossil record is shaped by sampling bias. Syst Biol 61:80–89
Lukoschek V, Keogh JS, Avise JC (2012) Evaluating fossil calibrations for dating phylogenies in light of rates of molecular evolution: a comparison of three approaches. Syst Biol 61:22–43
Lupia R, Lidgard S, Crane PR (1999) Comparing palynological abundance and diversity: implications for biotic replacement during the Cretaceous angiosperm radiation. Paleobiol 25:303–340
Magallon SA (2010) Using fossils to break long branches in molecular dating: a comparison of relaxed clocks allied to the origin of angiosperms. Syst Biol 59:384–399
Marshall CR (2008) A simple method for bracketing absolute divergence times on molecular phylogenies using multiple fossil calibration points. Amer Naturalist 171:726–742
McCormack JE, Heled J, Delaney KS, Peterson AT, Knowles LL (2010) Calibrating divergence times on species trees versus gene trees: implications for speciation history of Aphelocoma jays. Evolution 65:184–202
Nagalingum NS, Marshall CR, Quental TB, Rai HS, Little DP, Mathews S (2011) Recent synchronous radiation of a living fossil. Science 334:796–799
Near TJ, Sanderson MJ (2004) Assessing the quality of molecular divergence time estimates by fossil calibrations and fossil-based model selection. Philos Trans, Ser B 359:1477–1483
Niklas KJ, Tiffney BH, Knoll AH (1983) Patterns in vascular land plant diversification. Nature 303:614–616
Parham JF, Donoghue PCJ, Bell CJ, Calway TD, Head JJ, Holroyd PA, Inoue JG, Irmis RB, Joyce WG, Ksepka DT, Patané JSL, Smith ND, Tarver JE, van Tuinen M, Yang Z, Angielczyk KD, Greenwood JM, Hipsley CA, Jacobs L, Makovicky PJ, Müller J, Smith KT, Theodor JM, Warnock RCM, Benton MJ (2012) Best practices for justifying fossil calibrations. Syst Biol 61:346–359
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256 (http://darwin.uvigo.es/software/jmodeltest.html)
Pyron RA (2010) A likelihood method for assessing molecular divergence time estimates and the placement of fossil calibrations. Syst Biol 59:185–195
Qiu Y-L, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, Estabrook GF, Hendry TA, Taylor DW, Testa CM, Ambros M, Crandall-Stotler B, Duff RJ, Stech M, Frey W, Quandt D, Davis CC (2006) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA 103:15511–15516
Rabovsky DL (2010) Extintion rates should not be estimated from molecular phylogenies. Evolution 64:1816–1824
Rambaut A (2006–2009) FigTree. Tree figure drawing tool version 1.3.1, Institute of Evolutionary Biology, University of Edinburgh (http://tree.bio.ed.ac.uk/software/figtree/)
Rambaut A, Drummond AJ (2003–2009) Tracer. MCMC Trace analysis tool version v1.5.0 (http://tree.bio.ed.ac.uk/software/tracer/)
Raven P, Axelrod DI (1974) Angiosperm biogeography and past continental movements. Ann Missouri Bot Gard 61:539–673
Renner SS (2005) Relaxed molecular clocks for dating historical plant dispersal events. Trends Pl Sci 10:550–558
Rutschmann F, Eriksson T, Abu Salim K, Conti E (2007) Assessing calibration uncertainty in molecular dating: the assignments of fossils to alternative calibration points. Syst Biol 56:591–608
Sauquet H, Ho SYW, Gandolfo MA, Jordan GJ, Wilf P, Cantrill DJ, Bayly MJ, Bromham L, Brown GK, Carpenter RJ, Lee DM, Murphy DJ, Sniderman JMK, Udovicic F (2012) Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Syst Biol 61:289–313
Schneider H (2007) Plant morphology as the cornerstone to the integration of fossils and extant taxa in phylogenetic analyses. Sp Phyl Evol 1:65–71
Schneider H, Kenrick P (2001) An early Cretaceous root-climbing epiphyte (Lindsaeaceae) and its significance for calibrating the diversification of polypodiaceous ferns. Rev Palaeobot Palynol 115:33–41
Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallon S, Lupia R (2004) Ferns diversified in the shadow of angiosperms. Nature 428:553–557
Schneider H, Smith AR, Pryer KM (2009) Is morphology really at odds with molecules in estimating fern phylogeny. Syst Bot 34:455–475
Schneider H, Janssen T, Bystriakova N, Heinrichs J, Hennequin S, Rakotondrainibe F (2010a) Rapid radiations and neoendemism in the Madagascan biodiversity hotspot. In: Glaubrecht M (ed) Evolution in action: case studies in adaptive radiation, speciation, and the origin of biodiversity. Springer, Berlin, pp 3–15
Schneider H, Kreier H-P, Janssen T, Otto E, Muth H, Heinrichs J (2010b) Key innovations versus key opportunities; identifying causes of rapid radiations in derived ferns. In: Glaubrecht M (ed) Evolution in action: case studies in adaptive radiation, speciation, and the origin of biodiversity. Springer, Berlin, pp 61–76
Schuettpelz E, Pryer KM (2009) Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc Natl Acad Sci USA 27:11200–11205
Shaw AJ, Devos N, Cox CJ, Boles SB, Shaw B, Buchanan AM, Cave L, Seppelt R (2010) Peatmoss (Sphagnum) diversification associated with Miocene northern hemisphere climatic cooling? Molec Phylogen Evol 55:1139–1145
Smith AB (2001) Large-scale heterogeneity of the fossil record: implications for Phanerozoic biodiversity studies. Philos Trans, Ser B 356:351–367
Smith SA, Beaulieu JM, Stamatakis A, Donoghue MJ (2011) Understanding angiosperm diversification using small and large phylogenetic trees. Amer J Bot 98:404–414
Standke G (1998) Die Tertiärprofile der Samländischen Bernsteinküste bei Rauschen. Schriftenreihe Geowiss 7:93–133
Steenbock CM, Stockey RA, Beard G, Tomescu AMF (2011) A new family of leafy liverworts from the middle Eocene of Vancouver Island, British Columbia, Canada. Amer J Bot 98:998–1006
Tarver JE, Donoghue PCJ (2011) The trouble with topology: phylogenies without fossils provide a revisionist perspective of evolutionary history in topological analyses of diversity. Syst Biol 60:700–712
Taylor TN, Taylor EL, Krings M (2009) Paleobotany: the biology and evolution of fossil plants, 2nd edn. Academic Press, Boston
Vilnet AA, Konstantinova NA, Troitsky AV (2010) Molecular insight on phylogeny and systematics of the Lophoziaceae, Scapaniaceae, Gymnomitriaceae and Jungermanniaceae. Arctoa 19:31–50
Wagner PJ (2000) Exhaustion of morphological character states among fossil taxa. Evolution 54:365–386
Wang L, Schneider H, Zhang XC, Xiang QP (2012) The rise of the Himalaya enforced the diversification of SE Asian ferns by altering the monsoon regimes. BMC Plant Biol 12:210
Wilkinson RD, Steiper ME, Soligo C, Martin RD, Yang ZH, Tavaré S (2011) Dating primate divergences through an integrated analysis of palaeontological and molecular data. Syst Biol 60:16–31
Wilson R, Heinrichs J, Hentschel J, Gradstein SR, Schneider H (2007) Steady diversification of derived liverworts under Tertiary climatic fluctuations. Biol Lett 3:566–569
Yang XJ, Wu XW (2010) Sinolejeunea yimaensis gen. et. sp. nov. (Hepaticae) from the Middle-Jurassic Yima formation in Henan province of China. Sci China, Earth Sci 54:228–232
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. GARLI version 0.96 beta available online at http://www.nescent.org/informatics/
Acknowledgments
This study was supported by the German Research Foundation (Grant HE 3584/4) and funds from the estate of the late Dr. Ursula Hofmann, former lecturer at the Georg-August-University Göttingen. JV acknowledges support from the Ministry of Education of the Czech Republic through project No. 0021620828. HS acknowledges the Senior Visiting Professorship awarded by the Chinese Academy of Sciences. This is publication number 87 from the Courant Research Centre Geobiology that is funded by the German Initiative of Excellence.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Feldberg, K., Heinrichs, J., Schmidt, A.R. et al. Exploring the impact of fossil constraints on the divergence time estimates of derived liverworts. Plant Syst Evol 299, 585–601 (2013). https://doi.org/10.1007/s00606-012-0745-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-012-0745-y