Abstract
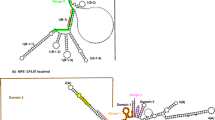
The primary and secondary structures of the intergenic spacer (IGS) between the 3′-end of 25S ribosomal RNA (rRNA) gene and the 5′-end of 18S rRNA gene are described for the cultivated chili pepper Capsicum pubescens. The recognized functional IGS is 2,078 bp in length. According to nucleotide base composition, regulatory elements, and conserved and repeated sequences the IGS can be divided into seven structural regions (SRI–VII). SRI comprises three copies of GAGGTTTTT-like motif, a probable transcription termination site in Solanaceae. At 3′-end, there are 21 bp matching the 18S rDNA. SRII is formed by 47 repeats of CACCATGG-like motif, the shortest repetitive region found in plant rDNA to date. SRIII is highly AT-rich, preceding SRIV, a highly conserved region in Solanaceae containing the transcription initiation site (TIS) TATATAAGGGGGG. The external transcribed spacer (ETS) is 966 bp in length. SRV-VII, downstream of the TIS, possesses eight inverted repeats, and three predicted stem-loops show pre-micro RNA (miRNA)-like structural features. Intragenomic variation is presented, and data are compared with characterized Solanaceae 25S–18S rDNA IGS.






Similar content being viewed by others
References
Backman TW, Sullivan CM, Cumbie JS, Miller ZA, Chapman EJ, Fahlgren N, Givan SA, Carrington JC, Kasschau KD (2008) Update of ASRP: the Arabidopsis Small RNA Project database. Nucleic Acids Res 36:D982–D985
Baldridge GD, Dalton MW, Fallon AM (1992) Is higher-order structure conserved in eukaryotic ribosomal DNA intergenic spacers? J Molec Evol 35:514–523
Barker RF, Harberd NP, Jarvis MG, Flavell RB (1988) Structure and evolution of the intergenic region in a rDNA repeat union of wheat. J Molec Biol 201:1–17
Borisjuk N, Hemleben V (1993) Nucleotide sequence of S. tuberosum rDNA intergenic spacer. Pl Molec Biol 21:381–384
Borisjuk N, Borisjuk L, Petjuch G, Hemleben V (1994) Comparison of nuclear ribosomal RNA genes among Solanum species and other Solanaceae. Genome 37:271–279
Borisjuk NV, Davidjuk YM, Kostishin SS, Miroshnichenco GP, Velasco R, Hemleben V, Volkov RA (1997) Structural analysis of rDNA in the genus Nicotiana. Plant Mol Biol 35:655–660
Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJ, Matzke MA (2009) Stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J 28:48–57
Delcasso-Treymousaygue D, Grellet F, Panabieres F, Ananiev E, Delseny M (1988) Structural and transcripcional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur J Biochem 172:767–776
Gruendler P, Unfried K, Pascher K, Schweizer D (1991) rDNA intergenic region from Arabidopsis thaliana: structural analysis, intraspecific variation and functional implications. J Molec Biol 221:1209–1222
Grundhoff A, Sullivan CS, Ganem D (2006) A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750
Hemleben V, Zentgraf U (1994) Structural organization and regulation of transcription by RNA polymerase I of plant nuclear ribosomal RNA genes. In: Nover L (ed) Results and problems in cell differentiation 20: plants promoters and transcription factors. Springer, Berlin/Heidelberg, pp 3–24
Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z (2007) MiPred: calssification of real and pseudo microRNA precursors using random forest prediciton model with combined features. Nucleic Acid Res 35:W339–W344
Kelly R, Siegel A (1989) The cucurbita maxima ribosomal DNA intergenic spacer has a complex structure. Gene 80:239–248
Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA (2004) Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol Biol 56:439–463
Komarova NY, Grimm GW, Hemleben V, Volkov RA (2008) Molecular evolution of 35S rDNA and taxonomic status of Lycopersicon within Solanum sect. Petota. Plant Syst Evol 276:59–71
Lafontaine D, Tollervey D (2001) Ribosomal RNA Encyclopedia of life sciences. Wiley, Chichester
McStay B, Grummt I (2008) The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24:131–157
Markham NR, Zuker M (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33:W577–W581
Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybriziation. In: Keith JM (ed) Bioinformatics, volume II structure, functions and applications, number 453 in methods in molecular biology, chapter 1. Humana, Totowa, pp 3–31
Martins TR, Barkman TJ (2005) Reconstruction of Solanaceae phylogeny using the nuclear gene SAMT. Syst Bot 30:435–447
Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21:367–376
Mayer C, Schmitz K-M, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22:351–361
Perry KL, Palukaitis P (1990) Transcription of S. lycopersicum ribosomal DNA and the organization of the intergenic spacer. Molec Gen Genet 221:102–112
Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, Pikaard CS (2008) Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell 32:673–684
Ricci A, Scali V, Passamonti M (2008) The IGS-ETS in Bacillus (Insecta Phasmida): molecular characterization and the relevance of sex in ribosomal DNA evolution. BMC Evol Biol 8:278
Ritchie W, Legendre M, Gautheret D (2007) RNA stem-loops: to be or not to be cleaved by RNAse III. RNA 13:457–462
Rogers SO, Bendich AJ (1987) Ribosomal RNA genes in plants: variability in copy number and in intergenic spacer. Plant Mol Biol 9:509–520
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plant, algae and fungi In: Stanton BG, Schilperoort RA (eds) A plant molecular biololgy manual. Kluwer Academic Publ, Dordrecht, pp D1/1-8
Schmidt-Puchta W, Guenther I, Saenger HL (1989) Nucleotide sequence of the intergenic spacer (IGS) of the S. lycopersicum ribosomal DNA. Plant Mol Biol 13:251–253
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tucker S, Vitins A, Pikaard C (2010) Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol 22:351–356
Volkov R, Kostishin S, Ehrendorfer F, Schweizer D (1996) Molecular organization and evolution of the external transcribed rDNA spacer region in two diploid relatives of Nicotiana tabacum (Solanaceae). Plant Syst Evol 201:117–129
Volkov RA, Bachmair A, Panchuk II, Kostyshyn SS, Schweizer D (1999a) 25S–18S rDNA intergenic spacer of Nicotiana sylvestris (Solanaceae): primary and secondary structure analysis. Plant Syst Evol 218:89–97
Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V (1999b) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol 16:311–320
Volkov RA, Komarova NY, Panchuk II, Hemleben V (2003) Molecular evolution of rDNA external transcribed spacer and phylogeny of sect. Petota (genus Solanum). Molec Phyl Evol 29:187–202
Volkov RA, Komarova NY, Hemleben (2007) Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Syst Biod 5:261–276
Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y (2005) MicroRNA identification based on sequence and structure alignment. Bioinformatics 21:3610–3614
Yousef M, Nebozhyn M, Shatkay H, Kanterakis S, Showe LC, Showe MK (2006) Combining multi-species genomic data for microRNA identification using a Naïve Bayes classifier. Bioinformatics 22:1325–1334
Zentgraf U, Hemleben V (1992) Complex formation of nuclear proteins with the RNA polymerase I promoter and repeated elements in the external transcribed spacer of Cucumis sativus ribosomal DNA. Nucl Acids Res 20:3685–3691
Acknowledgments
This study was supported by grant no. PICT 20196 of the Agencia Nacional de Promoción Científica y Tecnológica de Argentina (ANPCyT) and Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (CONICET) as part of the doctoral thesis of M.G. Additional support came from doctoral and postdoctoral research fellowships of CONICET to H.J.D. and M.G., respectively, who contributed equally to this work. H.J.D. carried out the DNA methods with the aid of M.G. and both performed the bioinformatic and secondary structures analyses. M.G. generated the figures and wrote the manuscript, and H.J.D., D.A.D., and E.A.M. revised it. The authors thank Prof. Elba Villanueva for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grabiele, M., Debat, H.J., Moscone, E.A. et al. 25S–18S rDNA IGS of Capsicum: molecular structure and comparison. Plant Syst Evol 298, 313–321 (2012). https://doi.org/10.1007/s00606-011-0546-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-011-0546-8