Abstract
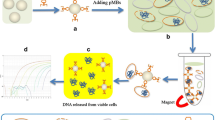
A colorimetric detection method for Escherichia coli (E. coli) in water was established based on a T7 phage tail fiber protein-magnetic separation. Firstly, the tail fiber protein (TFP) was expressed and purified to specifically recognize E. coli, which was verified by using fusion protein GFP-tagged TFP (GFP-TFP) and fluorescence microscopy. Then TFP conjugated with magnetic beads were applied to capture and separate E. coli. The TFP was covalently immobilized on the surface of magnetic beads and captured E. coli as verified by scanning electron microscopy (SEM). Finally, polymyxin B was used to lyse E. coli in solution and the released intracellular β-galactosidase (β-gal) could hydrolyze the colorimetric substrate chlorophenol red-β-D-galactopyranoside (CPRG), causing color change from yellow to purple. The high capture efficiencies of E. coli ranged from 88.70% to 95.65% and E. coli could be detected at a concentration of 102 CFU/mL by naked eyes. The specificity of the chromogenic substrate was evaluated using five different pathogen strains as competitors and tests with four kinds of real water samples showed recoveries of 86.00% to 92.25%. The colorimetric changes determined by visual inspection can be developed as an efficient platform for point-of-care detection of E. coli in resource-limited regions.
Graphical Abstract






Similar content being viewed by others
Data availability
Data will be made available on request.
References
Qi WZ, Zheng LY, Hou Y, Duan H, Wang L, Wang SY, Liu YJ, Li YB, Liao M, Lin JH (2022) A finger-actuated microfluidic biosensor for colorimetric detection of foodborne pathogens. Food Chem 381:131801. https://doi.org/10.1016/j.foodchem.2021.131801
Ripolles-Avila C, Martinez-Garcia M, Capellas M, Yuste J, Fung DYC, Rodriguez-Jerez JJ (2020) From hazard analysis to risk control using rapid methods in microbiology: A practical approach for the food industry. Compr Rev Food Sci Food Safety 19(4):1877–1907. https://doi.org/10.1111/1541-4337.12592
Wang DH, Chen JH, Nugen SR (2017) Electrochemical Detection of Escherichia coil from Aqueous Samples Using Engineered Phages. Anal Chem 89(3):1650–1657. https://doi.org/10.1021/acs.analchem.6b03752
Chen JH, Alcaine SD, Jiang ZW, Rotello VM, Nugen SR (2015) Detection of Escherichia coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal Chem 87(17):8977–8984. https://doi.org/10.1021/acs.analchem.5b02175
Mei JY, Wang DD, Zhang YH, Wu D, Cui JH, Gan MZ, Liu PF (2023) Portable paper-based nucleic acid enrichment for field testing. Adv Sci 10(11):e2205217. https://doi.org/10.1002/advs.202205217
Kulkarni MB, Ayachit NH, Aminabhavi TM (2023) Recent Advances in Microfluidics-Based Electrochemical Sensors for Foodborne Pathogen Detection. Biosens-Basel 13(2). https://doi.org/10.3390/bios13020246
Jia ZY, Muller M, Le Gall T, Riool M, Muller M, Zaat SAJ, Montier T, Schonherr H (2021) Multiplexed detection and differentiation of bacterial enzymes and bacteria by color-encoded sensor hydrogels. Bioact Mater 6(12):4286–4300. https://doi.org/10.1016/j.bioactmat.2021.04.022
Zhang LS, Huang R, Liu WP, Liu HX, Zhou XM, Xing D (2016) Rapid and visual detection of Listeria monocytogenes based on nanoparticle cluster catalyzed signal amplification. Biosens Bioelectron 86:1–7. https://doi.org/10.1016/j.bios.2016.05.100
Mannoor MS, Zhang SY, Link AJ, McAlpine MC (2010) Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc Natl Acad Sci USA 107(45):19207–19212. https://doi.org/10.1073/pnas.1008768107
Islam MA, Karim A, Ethiraj B, Raihan T, Kadier A (2022) Antimicrobial peptides: promising alternatives over conventional capture ligands for biosensor-based detection of pathogenic bacteria. Biotechnol Adv 55:107901. https://doi.org/10.1016/j.biotechadv.2021.107901
El-Boubbou K, Gruden C, Huang X (2007) Magnetic glyco-nanoparticles: A unique tool for rapid pathogen detection, decontamination, and strain differentiation. J Am Chem Soc 129(44):13392-+. https://doi.org/10.1021/ja076086e
Wang ZL, Cai R, Gao ZP, Yuan YH, Yue TL (2020) Immunomagnetic separation: An effective pretreatment technology for isolation and enrichment in food microorganisms detection. Compr Rev Food Sci Food Safety 19(6):3802–3824. https://doi.org/10.1111/1541-4337.12656
Srisa-Art M, Boehle KE, Geiss BJ, Henry CS (2018) Highly Sensitive Detection of Salmonella typhimurium Using a Colorimetric Paper-Based Analytical Device Coupled with Immunomagnetic Separation. Anal Chem 90(1):1035–1043. https://doi.org/10.1021/acs.analchem.7b04628
Liu MY, Yue FL, Kong QQ, Liu ZL, Guo YM, Sun X (2022) Aptamers against Pathogenic Bacteria: Selection Strategies and Apta-assay/Aptasensor Application for Food Safety. J Agric Food Chem 70(18):5477–5498. https://doi.org/10.1021/acs.jafc.2c01547
Zhang WS, Cui CJ, Chen HS, Liu HB, Bin S, Wang DL, Wang YT (2022) Advances in electrochemical aptamer biosensors for the detection of food-borne pathogenic bacteria. ChemistrySelect 7(29):e202202190. https://doi.org/10.1002/slct.202202190
Zhang XR, Chen LL, Hu SW (2021) Advances in Bacteria Biosensing Based on Molecular Recognition. Chem J Chinese Univ-Chinese 42(11):3468–3476. https://doi.org/10.7503/cjcu20210425
Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, Dutilh BE, Brouns SJJ (2018) Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16(12):760–773. https://doi.org/10.1038/s41579-018-0070-8
Silva JB, Storms Z, Sauvageau D (2016) Host receptors for bacteriophage adsorption. Fems Microbiol Lett 363(4):fnw002. https://doi.org/10.1093/femsle/fnw002
Costa SP, Nogueira CL, Cunha AP, Lisac A, Carvalho CM (2022) Potential of bacteriophage proteins as recognition molecules for pathogen detection. Crit Rev Biotechnol 18:1–18. https://doi.org/10.1080/07388551.2022.2071671
He Y, Shi YL, Liu ML, Wang YR, Wang L, Lu SG, Fu ZF (2018) Nonlytic Recombinant Phage Tail Fiber Protein for Specific Recognition of Pseudomonas aeruginosa. Anal Chem 90(24):14462–14468. https://doi.org/10.1021/acs.analchem.8b04160
Bai YL, Shahed-Al-Mahmud M, Selvaprakash K, Lin NT, Chen YC (2019) Tail Fiber Protein-Immobilized Magnetic Nanoparticle-Based Affinity Approaches for Detection of Acinetobacter baumannii. Anal Chem 91(15):10335–10342. https://doi.org/10.1021/acs.analchem.9b02964
Chen Y, Yang HL, Luo S, Wang L, Lu SG, Fu ZF (2022) Engineering phage tail fiber protein as a wide-spectrum probe for Acinetobacter baumannii strains with a recognition rate of 100%. Anal Chem 94(27):9610–9617. https://doi.org/10.1021/acs.analchem.2c00682
Cerritelli ME, Conway JF, Cheng NQ, Trus BL, Steven AC (2003) Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem 64:301–323. https://doi.org/10.1016/s0065-3233(03)01008-8
Garcia-Doval C, van Raaij MJ (2012) Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc Natl Acad Sci USA 109(24):9390–9395. https://doi.org/10.1073/pnas.1119719109
Dunne M, Rupf B, Tala M, Qabrati X, Ernst P, Shen Y, Sumrall E, Heeb L, Pluckthun A, Loessner MJ, Kilcher S (2019) Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Reports 29(5):1336-+. https://doi.org/10.1016/j.celrep.2019.09.062
Wang M, Deng ZF, Li YM, Ma Y, Wang JF (2022) Design and characterization of a novel lytic protein against Clostridium difficile. Appl Microbiol Biotechnol 106(12):4511–4521. https://doi.org/10.1007/s00253-022-12010-0
Witte S, Zinsli LV, Gonzalez-Serrano R, Matter CI, Loessner MJ, van Mierlo JT, Dunne M (2021) Structural and functional characterization of the receptor binding proteins of Escherichia coli O157 phages EP75 and EP335. Comput Struct Biotechnol J 19:3416–3426. https://doi.org/10.1016/j.csbj.2021.06.001
Santos SB, Cunha AP, Macedo M, Nogueira CL, Brandao A, Costa SP, Melo LDR, Azeredo J, Carvalho CM (2020) Bacteriophage-receptor binding proteins for multiplex detection ofStaphylococcusandEnterococcusin blood. Biotechnol Bioeng 117(11):3286–3298. https://doi.org/10.1002/bit.27489
Xiao FB, Li WQ, Xu HY (2022) Advances in magnetic nanoparticles for the separation of foodborne pathogens: Recognition, separation strategy, and application. Compr Rev Food Sci Food Safety. https://doi.org/10.1111/1541-4337.13023
Filik K, Szermer-Olearnik B, Oleksy S, Brykala J, Brzozowska E (2022) Bacteriophage tail proteins as a tool for bacterial pathogen recognition-a literature review. Antibiot-Basel 11(5):555. https://doi.org/10.3390/antibiotics11050555
Mao Y, Huang XL, Xiong SC, Xu HY, Aguilar ZP, Xiong YH (2016) Large-volume immunomagnetic separation combined with multiplex PCR assay for simultaneous detection of Listeria monocytogenes and Listeria ivanovii in lettuce. Food Control 59:601–608. https://doi.org/10.1016/j.foodcont.2015.06.048
Bayramoglu G, Ozalp VC, Oztekin M, Arica MY (2019) Rapid and label-free detection of Brucella melitensis in milk and milk products using an aptasensor. Talanta 200:263–271. https://doi.org/10.1016/j.talanta.2019.03.048
Dursun AD, Borsa BA, Bayramoglu G, Arica MY, Ozalp VC (2022) Surface plasmon resonance aptasensor for Brucella detection in milk. Talanta 239:123074. https://doi.org/10.1016/j.talanta.2021.123074
Zhou Y, Ramasamy RP (2019) Isolation and separation of Listeria monocytogenes using bacteriophage P100-modified magnetic particles. Colloids Surf B-Biointerfaces 175:421–427. https://doi.org/10.1016/j.colsurfb.2018.12.007
Wang ZY, Wang DH, Kinchla A, Sela D, Nugen S (2016) Rapid screening of waterborne pathogens using phage-mediated separation coupled with real-time PCR detection. Anal Bioanal Chem 408(15):4169–4178. https://doi.org/10.1007/s00216-016-9511-2
Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S (2014) Structure of beta-galactosidase at 3.2-angstrom resolution obtained by cryo-electron microscopy. Proceed Natl Acad Sci US A 111(32):11709–11714. https://doi.org/10.1073/pnas.1402809111
Sun QL, Cao MC, Zhang X, Wang M, Ma Y, Wang JF (2021) A simple and low-cost paper-based colorimetric method for detecting and distinguishing the GII.4 and GII.17 genotypes of norovirus. Talanta 225:121978. https://doi.org/10.1016/j.talanta.2020.121978
Denyes JM, Dunne M, Steiner S, Mittelviefhaus M, Weiss A, Schmidt H, Klumpp J, Loessner MJ (2017) Modified bacteriophage S16 long tail fiber proteins for rapid and specific immobilization and detection of salmonella cells. Appl Environ Microbiol 83(12):e00277-17. https://doi.org/10.1128/aem.00277-17
Xu JZ, Li XB, Kang GB, Bai L, Wang P, Huang H (2020) Isolation and characterization of AbTJ, an Acinetobacter baumannii phage, and functional identification of its receptor-binding modules. Viruses-Basel 12(2):205. https://doi.org/10.3390/v12020205
Wang LK, Lin H, Zhang J, Wang JX (2022) Phage long tail fiber protein-immobilized magnetic nanoparticles for rapid and ultrasensitive detection of Salmonella. Talanta 248:123627. https://doi.org/10.1016/j.talanta.2022.123627
Ding YF, Huang CX, Zhang YM, Wang J, Wang XH (2023) Magnetic microbead enzyme-linked immunoassay based on phage encoded protein RBP 41-mediated for rapid and sensitive detection of Salmonella in food matrices. Food Res Int 163:112212. https://doi.org/10.1016/j.foodres.2022.112212
Acknowledgements
This work was supported by Science and Technology Program of Guangzhou (202205110007).
Author information
Authors and Affiliations
Contributions
Bin Hong, Yi Ma and Jufang Wang, designed the experiments. Bin Hong, wrote the manuscript and performed the experiments. Yanmei Li, provided writing assistance for the manuscript. Wenhai Wang, analyzed data and drafted the manuscript. All authors have approved and read the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
1.A colorimetric detection method for E. coli in water was established based on a T7 phage tail fiber protein-magnetic separation.
2.Polymyxin B showed good lytic ability and was used to lyse E. coli to release β-gal.
3.There are high separation rate and low limit of detection for E. coli in our study.
4.The colorimetric change can be determined by naked eyes allowing for point-of-care testing of E. coli in resource-limited regions.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hong, B., Li, Y., Wang, W. et al. Separation and colorimetric detection of Escherichia coli by phage tail fiber protein combined with nano-magnetic beads. Microchim Acta 190, 202 (2023). https://doi.org/10.1007/s00604-023-05784-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05784-1