Abstract
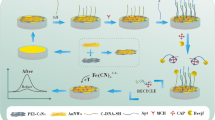
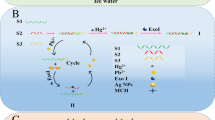
An electrochemiluminescence (ECL) DNA biosensor based on ExoIII exonuclease assistance and hybridization chain reaction (HCR) amplification technology has been constructed. ExoIII exonuclease and triple-helix DNA molecular switch are used in detecting a target in circulation. By combining HCR with AuNPs@DNA, a novel signal probe is built, which enables multiple signal amplification and the high-sensitive detection of transgenic rice BT63 DNA. The Fe3O4@Au solution is added to a magneto-controlled glassy carbon electrode, and sulfhydryl-modified capture DNA (CP) is immobilized on Fe3O4@Au through the Au–S bond. Mercaptoethanol is added to close sites and prevent the nonspecific adsorption of CP on the magnetron glassy carbon electrode. A target DNA is added to a constructed triple-helix DNA molecular centrifuge tube for reaction. Owing to base complementation and the reversible switching of the triple-helix DNA molecular state, the target DNA turns on the triple-helix DNA molecular switch and hybridizes with a long-strand recognition probe (RP) to form a double-stranded DNA (dsDNA). Exonuclease ExoIII is added to specifically recognize and cut the dsDNA and to release the target DNA. The target DNA strand then circulates back completely to open the multiple triple-helix DNA molecular switch, releasing a large number of signal transduction probes (STP). To hybridize with CP, a large amount of STP is added to the electrode. Finally, a AuNPs@DNA signal probe is added to hybridize with STP. H1 and H2 probes are added for the hybridization chain reaction and the indefinite extension of the primer strand on the probe. Then, tris-(bipyridyl)ruthenium(II) is added for ECL signal detection with PBS-tri-n-propylamine as the base solution. In the concentration range 1.0 × 10−16 to 1.0 × 10−8 mol/L of the target DNA, good linear relationship was achieved with the corresponding ECL signal. The detection limit is 3.6 × 10−17 mol/L. The spiked recovery of the rice samples range from 97.2 to 101.5%. The sensor is highly sensitive and has good selectivity, stability, and reproducibility.
Graphical abstract

A novel electrochemiluminescence biosensor with extremely higher sensitivity was prepared for the determination of ultra-trace amount transgenic rice BT63 DNA. The sensitivity was significantly improved by multiple signal enhancements. Firstly, a large number of signal transduction probes are released when the triple-helix DNA molecular switch unlock after recycles assisted by ExoIII exonuclease under target BT63 DNA; and then the signal transduction probes hybridize with the signal probes of AuNPs@(DNA-HCR) produced through hybridization chain reaction. Finally, the signal probes which were embedded with a large amount of electrochemiluminescence reagent produce high luminescence intensity. The detection limit was 3.6 × 10−17 mol/L, which is almost the most sensitive methods reported.






Similar content being viewed by others
References
He CM, Tang YL, Wang S, Liu JH, Chen Y, Dong YY, Su HJ, Tan TW (2017) An electrochemical DNA biosensor based on au-reduced graphene oxide nanocomposite for transgenic event Bt63 detection. Anal Sci 33:1155–1160
Cottenet G, Blancpain C, Sonnard V, Chuah PF (2019) Two FAST multiplex real-time PCR reactions to assess the presence of genetically modified organisms in food. Food Chem 274:760–765
Chen M, Zhao JZ, Ye GY, Fu Q, Shelton AM (2016) Impact of insect-resistant transgenic rice on target insect pests and non-target arthropods in China. Insect Sci 13:409–420
Xia H, Chen LY, Wang F, Lu BR (2010) Yield benefit and underlying cost of insect-resistance transgenic rice: implication in breeding and deploying transgenic crops. Field Crop Res 118:215–220
Gao DY, Sheng ZH, Han HY (2011) An ultrasensitive method for the detection of gene fragment from transgenics using label-free gold nanoparticle probe and dynamic light scattering. Anal Chim Acta 696:1–5
Ma PF, Ye H, Deng JY, Khan IM, Yue L, Wang ZP (2019) A fluorescence polarization aptasensor coupled with polymerase chain reaction and streptavidin for chloramphenicol detection. Talanta 205:120119
Ahmed MU, Saito M, Hossain MM, Rao SR, Furui S, Hino A, Takamura Y, Takagi M, Tamiya E (2009) Electrochemical genosensor for the rapid detection of GMO using loop-mediated isothermal amplification. Analyst 134:966–972
Rajagopal P, Chellappan DR, Sridharan S, Pemiah B, Krishnaswamy S, Sethuraman S, Sekar KR, Krishnan UM (2020) Microarray analysis of genes from animals treated with a traditional formulation ChandraprabhaVati reveals its therapeutic targets. J Tradit Complement Med 10:36–44
Chow YY, Rahman S, Ting ASY (2019) Evaluating the host defense responses in oil palm to complex biocontrol endophyte–pathogen–host plant interaction via Fluidigm® real-time polymerase chain reaction (RT-PCR). Biol Control 129:148–157
Sammons DL, Biagini RE, Smith JP, MacKenzie BA, Striley CAF, Roberston SA, Snawder JE, Ferguson BS, Larkin KA (2007) Simultaneous measurement of bacillus thuringiensis Cry1Ab and Cry3B proteins in corn extracts. ACS Sym Ser 966:274–285
Bulcke MVD, Schrijver AD, Bernardi DD, Devos Y, MbongoMbella G, Casi AL, Moens W, Sneyers M (2007) Detection of genetically modified plant products by protein strip testing: an evaluation of real-life samples. Eur Food Res Technol 225:49–57
Becherer L, Bakheit M, Frischmann S, Stinco S, Borst N, Zengerle R, Stetten FV (2018) Simplified real-time multiplex detection of loop-mediated isothermal amplification using novel mediator displacement probes with universal reporters. Anal Chem 90:4741–4748
Zhang M, Hai H, Zhou FY, Zhong JC, Li JP (2018) Electrochemical luminescent DNA sensor based on polymerase-assisted signal amplification. Chin J Anal Chem 46:203–209
Wang XJ, Niu SY, Wei MM, Liu S, Rui Liu R, Shi C, Ma CP (2020) Ultrasensitive electrochemical DNA biosensor based on a tetrahedral structure and proximity-dependent surface hybridization. Analyst 145:150–156
Wang LL, Wen YL, Yang X, Xu L, Liang W, Zhu Y, Wang LH, Li Y, Li Y, Ding M, Ren SZ, Yang ZZ, Lv M, Zhang JC, Ma K, Liu G (2019) Ultrasensitive electrochemical DNA biosensor based on a label-free assembling strategy using a triblock polyA DNA probe. Anal Chem 91(24):16002–16009
Chen M, Su HL, Mao L, Guo MY, Tang JL (2018) Highly sensitive electrochemical DNA sensor based on the use of three-dimensional nitrogen-doped graphene. Microchim Acta 185:51
Bonsu DOM, Denice H, Austin JJ (2020) Forensic touch DNA recovery from metal surfaces–a review. Sci Justice 60:206–215
Hegg EL, Burstyn JN (1996) Copper(II) macrocycles cleave dingle-stranded and double-stranded DNA under both aerobic and anaerobic conditions. Inorg Chem 35:7474–7481
Ramezani M, Danesh NM, Lavaee P, Abnouse K, Taghdisi SM (2015) A novel colorimetric triple-helix molecular switch aptasensor for ultrasensitive detection of tetracycline. Biosens Bioelectron 70:181–187
Bagheri E, Abnous K, Alibolandi M, Ramezani M, Taghdisi SM (2018) Triple-helix molecular switch-based aptasensors and DNA sensors. Biosens Bioelectron 111:1–9
Sun H, Yao FJ, Su ZQ, Kang XF (2020) Hybridization chain reaction (HCR) for amplifying nanopore signals. Biosens Bioelectron 150:111906
Li XY, Li JJ, Zhu CX, Zhang XH, Chen JH (2018) A new electrochemical immunoassay for prion protein based on hybridization chain reaction with hemin/G-quadruplex DNAzyme. Talanta 182:292–298
Zhang B, Liu BQ, Tang DP, Niessner R, Chen G, Knopp D (2012) DNA-based hybridization chain reaction for amplified bioelectronic signal and ultrasensitive detection of proteins. Anal Chem 84(12):5392–5399
Zhang KY, Lv SZ, Zhou Q, Tang DP (2020) CoOOH nanosheets-coated g-C3N4/CuInS2 nanohybrids for photoelectrochemical biosensor of carcinoembryonic antigen coupling hybridization chain reaction with etching reaction. Sensors Actuators B Chem 307:127631
Zeng RJ, Zhang LJ, Su LS, Luo ZB, Zhou Q, Tang DP (2019) Photoelectrochemical bioanalysis of antibiotics on rGO-Bi2WO6-au based on branched hybridization chain reaction. Biosens Bioelectron 133:100–106
Gao ZQ, Qiu ZL, Lu MH, Shu J, Tang DP (2017) Hybridization chain reaction-based colorimetric aptasensor of adenosine 5′-triphosphate on unmodified gold nanoparticles and two label-free hairpin probes. Biosens Bioelectron 89:1006–1012
Cheng H, Liu JQ, Ma WJ, Duan SD, Huang J, He XX, Wang KM (2018) Low background cascade signal amplification electrochemical sensing platform for tumor-related mRNA quantification by target-activated hybridization chain reaction and electroactive cargo release. Anal Chem 90:12544–12552
Feng KJ, Liu J, Deng L, Yu HJ, Yang MH (2018) Amperometric detection of microRNA based on DNA-controlled current of a molybdophosphate redox probe and amplification via hybridization chain reaction. Microchim Acta 185:28
Yun W, Wu H, Chen L, Yang LZ (2018) Dual enzyme-free amplification strategy for ultra-sensitive fluorescent detection of bisphenol a in water. Anal Chim Acta 1020:104–109
Wang LP, Huang YB, Lai YH (2018) Surface enhanced Raman scattering activity of dual-functional Fe3O4/Au composites. Appl Surf Sci 435:290–296
Alaa A, Yazan A, Mazhar SZ, Khalid MB, Bahaa T, Osama AA, Alaaldin MA, Mourad B, David JE (2018) Synthesis of gold nanoparticles using leaf extract of ziziphus zizyphus and their antimicrobial activity. Nanomaterials 8:174–186
Zhang Y, Xu GY, Lian GL, Luo F, Xie QF, Lin ZY, Chen GN (2020) Electrochemiluminescence biosensor for miRNA-21 based on toehold-mediated strand displacement amplification with Ru(phen)32+ loaded DNA nanoclews as signal tags. Biosens Bioelectron 147:111789
Tang L, Zeng GM, Shen GL, Li YP, Liu C, Li Z, Luo J, Fan CZ, Yang CP (2009) Sensitive detection of lip genes by electrochemical DNA sensor and its application in polymerase chain reaction amplicons from Phanerochaete chrysosporium. Biosens Bioelectron 24:1474–1479
Sun W, Qin P, Gao HW, Li GC, Jiao K (2010) Electrochemical DNA biosensor based on chitosan/nano-V2O5/MWCNTs composite film modified carbon ionic liquid electrode and its application to the LAMP product of Yersinia enterocolitica gene sequence. Biosens Bioelectron 25:1264–1270
Knez K, Spasic D, Delport F, Lammertyn J (2015) Real-time ligation chain reaction for DNA quantification and identification on the FO-SPR. Biosens Bioelectron 67:394–399
Acknowledgments
The authors gratefully acknowledge the financial support received from the National Nature Science Foundation of China (21765006), Natural Science Foundation of Guangxi Province of China (2018GXNSFAA050023), and the Research Program of Guangxi Specially-Invited Experts (key technologies for intensive processing and quality safety of agricultural products, TingFa[2018]39).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 65 kb)
Rights and permissions
About this article
Cite this article
Hai, H., Chen, C., Chen, D. et al. A sensitive electrochemiluminescence DNA biosensor based on the signal amplification of ExoIII enzyme-assisted hybridization chain reaction combined with nanoparticle-loaded multiple probes. Microchim Acta 188, 125 (2021). https://doi.org/10.1007/s00604-021-04777-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04777-2