Abstract
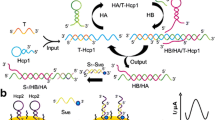
Voltammetric detection of the K-ras gene fragment was accomplished through the combined application of (a) a switchable DNA nanostructure, (b) the use of hairpin probe and exonuclease III (Exo III)-assisted signal amplification, (c) a split G-quadruplex, and (d) by exploiting the redox activity of DNAzyme. Three assistant oligonucleotides were designed to construct a DNA tweezer on a gold electrode. It is in “open state” in the absence of K-ras DNA. Then, a hairpin probe was introduced, whose stem-loop structure can be opened through hybridization with the K-ras DNA. Exo III is added which hydrolyzes the complementary region of the hairpin sequence to release a single-stranded rest fragment. The ssDNA hybridizes with the DNA tweezer on the electrode which thereby is switched to the “closed state”. This leads to the formation of G-quadruplex due to the shortened distance of the split G-quadruplex-forming sequences in the tweezer. The voltammetric signal of the G-quadruplex-hemin complex, with a peak near −0.3 V vs. Ag/AgCl, is used as the signal output. Under the optimal conditions, the current response in differential pulse voltammetry (DPV) increases linearly with the concentration of K-ras DNA in the range of 0.01–1000 pM, and the detection limit is 2.4 fM. The assay can clearly discriminate K-ras DNA from a single-base mutation. The method has excellent selectivity and was applied to the determination of K-ras DNA in (spiked) serum samples.

Schematic representation of a method for the determination of the K-ras gene fragment through a combination of switchable DNA tweezer, split G-quadruplex, and exonuclease III (ExoIII)-assisted target recycling signal amplification.







Similar content being viewed by others
References
Zhang G, Tang Z, Wang W, Wu H, Maham A, Lin Y (2010) Hairpin DNA switch for ultrasensitive spectrophotometric detection of DNA hybridization based on gold nanoparticles and enzyme signal amplification. Anal Chem 82:6440–6446. https://doi.org/10.1021/ac1006238
Liu Z, Zhang W, Zhu S, Zhang L, Hu L, Parveen S, Xu G (2011) Ultrasensitive signal-on DNA biosensor based on nicking endonuclease assisted electrochemistry signal amplification. Biosens Bioelectron 29:215–218. https://doi.org/10.1016/j.bios.2011.07.076
Jing Z, Ding Y, Liu X, Wang L, Jiang W (2014) Toehold-mediated strand displacement reaction triggered isothermal DNA amplification for highly sensitive and selective fluorescent detection of single-base mutation. Biosens Bioelectron 59:276–281. https://doi.org/10.1016/j.bios.2014.03.051
Wei W, Ni Q, Pu Y, Yin L, Liu S (2014) Electrochemical biosensor for DNA damage detection based on exonuclease III digestions. J Electroanal Chem 714:25–29. https://doi.org/10.1016/j.jelechem.2013.12.018
Wang H, Ma R, Sun F, Jia L, Zhang W, Shang L, Xue Q, Jia W, Wang H (2018) A versatile label-free electrochemical biosensor for circulating tumor DNA based on dual enzyme assisted multiple amplification strategy. Biosens Bioelectron 122:224–230. https://doi.org/10.1016/j.bios.2018.09.028
Lei G, Sun X, Hong Q, Li F (2017) Ratiometric NanoCluster Beacon: a label-free and sensitive fluorescent DNA detection platform. ACS Appl Mater Interf 9:13102–13110. https://doi.org/10.1021/acsami.7b03198
Ge L, Wang W, Li F (2017) Electro-grafted electrode with graphene-oxide-like DNA affinity for ratiometric homogeneous electrochemical biosensing of microRNA. Anal Chem 89:11560–11567. https://doi.org/10.1021/acs.analchem.7b02896
Li Y, Miao X, Ling L (2015) Triplex DNA: a new platform for polymerase chain reaction – based biosensor. Sci Rep 5:13010. https://doi.org/10.1038/srep13010
Chen Q, Bian Z, Chen M, Hua X, Yao C, Xia H, Kuang H, Zhang X, Huang J, Cai G (2009) Real-time monitoring of the strand displacement amplification (SDA) of human cytomegalovirus by a new SDA-piezoelectric DNA sensor system. Biosens Bioelectron 24:3412–3418. https://doi.org/10.1016/j.bios.2009.06.012
Zhang Y, Wang L, Luo F, Qiu B, Guo L, Weng Z, Lin Z, Chen G (2017) An electrochemiluminescence biosensor for Kras mutations based on locked nucleic acid functionalized DNA walkers and hyperbranched rolling circle amplification. Chem Comm 53:2910–2913. https://doi.org/10.1039/c7cc00009j
Sun X, Chen H, Wang S, Zhang Y, Tian Y, Zhou N (2018) Electrochemical detection of sequence-specific DNA based on formation of G-quadruplex-hemin through continuous hybridization chain reaction. Anal Chim Acta 2012:121–128. https://doi.org/10.1016/j.aca.2018.02.076
Hou T, Li W, Liu X, Li F (2015) Label-free and enzyme-free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: a facile, sensitive, and highly specific microRNA assay. Anal Chem 87:11368–11374. https://doi.org/10.1021/acs.analchem.5b02790
Li S, Chen A, Chai Y, Yuan R, Zhuo Y (2016) Electrochemiluminescence aptasensor based on cascading amplification of nicking endonuclease-assisted target recycling and rolling circle amplifications for mucin 1 detection. Electrochim Acta 212:767–774. https://doi.org/10.1016/j.electacta.2016.07.074
Zhou W, Gong X, Xiang X, Yuan R, Chai Y (2014) Quadratic recycling amplification for label-free and sensitive visual detection of HIV DNA. Biosens Bioelectron 55:220–224. https://doi.org/10.1016/j.bios.2013.12.021
Wu H, Zhang K, Liu Y, Wang H, Wu J, Zhu F, Zou P (2015) Binding-induced and label-free colorimetric method for protein detection based on autonomous assembly of gemin/G-quadruplex DNAzyme amplification strategy. Biosens Bioelectron 64:572–578. https://doi.org/10.1016/j.bios.2014.09.096
Li H, Carter JD, Labean TH (2010) Nanofabrication by DNA self-assembly. Mater Today 12:24–32. https://doi.org/10.1016/S1369-7021(09)70157-9
Jonathan B, Turberfield AJ (2007) DNA nanomachines. Nature Nanotech 2:275–284. https://doi.org/10.1038/nnano.2007.104
Liu M, Fu J, Hejesen C, Yang Y, Woodbury N, Gothelf K, Liu Y, Yan H (2013) A DNA tweezer-actuated enzyme nanoreactor. Nat Commun 4:2127. https://doi.org/10.1038/ncomms3127
Gu H, Chao J, Xiao S, Seeman N (2010) A proximity-based programmable DNA nanoscale assembly line. Nature 465:202–205. https://doi.org/10.1038/nature09026
Kou B, Chai Y, Yuan Y, Yuan R (2018) Dynamical regulation of enzyme cascade amplification by a regenerated DNA nanotweezer for ultrasensitive electrochemical DNA detection. Anal Chem 90:10701–11076. https://doi.org/10.1021/acs.analchem.8b00477
Li D, Cheng W, Li Y, Li X, Yin X, Yin Y, Ju H, Ding S (2015) Catalytic hairpin assembly actuated DNA nanotweezer for logic gate building and sensitive enzyme-free biosensing of microRNAs. Anal Chem 88:7500–7506. https://doi.org/10.1021/acs.analchem.5b04844
Andersen E, Dong M, Nielsen M, Jahn K, Subramani R, Mamdouh W, Golas M, Sander B, Stark H, Oliveira C (2009) Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459:73–76. https://doi.org/10.1038/nature07971
Zhang H, Lai M, Zuehlke A, Peng H, Li X, Le X (2016) Binding-induced DNA nanomachines triggered by proteins and nucleic acids. Angew Chem Int Ed 127:14534–14538. https://doi.org/10.1002/anie.201506312
Zhang P, Li Z, Wang H, Zhuo Y, Yuan R, Chai Y (2017) DNA nanomachine-based regenerated sensing platform: a novel electrochemiluminescence resonance energy transfer strategy for ultra-high sensitive detection of microRNA from cancer cells. Nanoscale 9:2310–2316. https://doi.org/10.1039/c6nr08631d
Liu S, Su W, Li Z, Ding X (2015) Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures. Biosens Bioelectron 71:57–61. https://doi.org/10.1016/j.bios.2015.04.006
Xu Z, Liao L, Chai Y, Wang R, Yuan R (2017) Ultrasensitive electrochemiluminescence biosensor for microRNA detection by 3D DNA walking machine based target conversion and distance-controllable signal quenching and enhancing. Anal Chem 89:8282–8287. https://doi.org/10.1021/acs.analchem.7b01409
Xu Y, Luo X, Geng N, Wu M, Lu Z (2018) DNA nanotweezers with hydrolytic activity for enzyme-free and sensitive detection of fusion gene via logic operation. J Anal Methods Chem 2018:7. https://doi.org/10.1155/2018/4178045
Gong X, Zhou W, Li D, Chai Y, Xiang Y, Yuan R (2015) RNA-regulated molecular tweezers for sensitive fluorescent detection of microRNA from cancer cells. Biosens Bioelectron 71:98–102. https://doi.org/10.1016/j.bios.2015.04.003
Funabashi H, Shigeto H, Nakatsuka K, Kuroda A (2015) A FRET-based DNA nano-tweezer technique for the imaging analysis of specific mRNA. Analyst 140:999. https://doi.org/10.1039/c4an02064b
Bian X, Guo B, Zhao M, Han D, Cheng W, Song F, Ding S (2019) An enzyme-free “ON-OFF” electrochemiluminescence biosensor for ultrasensitive detection of PML/RARα based on target-switched DNA nanotweezer. ACS AMI 11:3715–3721. https://doi.org/10.1021/acsami.8b18497
Li D, Wei C, Li Y, Xu Y, Li X, Yin Y, Ju H, Ding S (2016) Catalytic hairpin assembly actuated DNA nanotweezer for logic gate building and sensitive enzyme-free biosensing of microRNAs. Anal Chem 88:7500–7506. https://doi.org/10.1021/acs.analchem.5b04844
Yoshikawa K, Tanabe E, Shibata A, Inoue S, Kitayoshi M, Okimoto S, Fukushima N, Tsujiuchi T (2013) Involvement of oncogenic K-ras on cell migration stimulated by lysophosphatidic acid receptor-2 in pancreatic cancer cells. Exp Cell Res 319:105–112. https://doi.org/10.1016/j.yexcr.2012.09.014
Sun X, Wang S, Zhang Y, Tian Y, Zhou N (2017) Ultrasensitive detection of DNA based on target-triggered hairpin assembly and exonuclease-assisted recycling amplification. Sens Actuat B-Chem 252:306–312. https://doi.org/10.1016/j.snb.2017.06.014
Lin L, Liu A, Zhao C, Weng S, Lei Y, Liu Q, Lin X, Chen Y (2013) A chronocoulometric LNA sensor for amplified detection of K-ras mutation based on site-specific DNA cleavage of restriction endonuclease. Biosens Bioelectron 42:409–414. https://doi.org/10.1016/j.bios.2012.09.063
Xu H, Wu D, Li C, Lu Z, Liao X, Huang J, Wu Z (2017) Label-free colorimetric detection of cancer related gene based on two-step amplification of molecular machine. Biosens Bioelectron 90:314–320. https://doi.org/10.1016/j.bios.2016.12.003
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 31271860), the Six Talent Peaks Project in Jiangsu Province (JY-078) and National First-class Discipline Program of Light Industry Technology and Engineering (LITE2018-07).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Ethical approval
All procedures were approved by medical ethics committee of Jiangnan University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, H., Sun, X., Cai, R. et al. Switchable DNA tweezer and G-quadruplex nanostructures for ultrasensitive voltammetric determination of the K-ras gene fragment. Microchim Acta 186, 843 (2019). https://doi.org/10.1007/s00604-019-3993-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3993-5