Abstract
A new fluorometric method is delineated for the detection of RNase H activity by combining DNAzyme with reduced graphene oxide (rGO). In the absence of RNase H, the fluorescence of FAM-labeled probe is quenched due to the strong adsorption on the rGO. The presence of RNase H can release the active DNAzyme from the DNA-RNA chimeric strand. This triggers the cleavage of the signal probe at the rA site with the help of the cofactor Mg2+. The recycle cleavage can directly result in the amplified signal emitted by the FAM-labeled short fragment. The method allows the activity of RNase H to be detected in a linear range of 0.01 to 5 U·mL−1. The detection limit of 0.018 U·mL−1 is calculated by the principle of three-time standard deviation over the blank signal. Then, RNase H-targeting natural compounds were screened for their inhibitory action. Among the investigated compounds, five were screened as RNase H inhibitors in a concentration-dependent manner, and 4 compounds were identified as activators. Finally, the method was reliably used for discriminating the difference of RNase H activity in human serum. It is found that RNase H activity was upregulated in patients with hepatitis C virus infection.

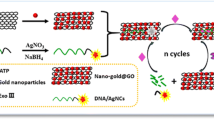
The schematic presentation of rGO-DNAzyme-based RNase H detection. RNase H triggers the active DNAzyme releasing from the DNA-RNA chimeric strand, which can cleavage probes to FAM-labeled short fragments and make the fluorescence signal cycle amplified.






Similar content being viewed by others
References
Cerritelli SM, Crouch RJ (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J 276(6):1494–1505
Tadokoro T, Kanaya S (2009) Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J 276(6):1482–1493
Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121(7):1005–1016
Kiełpiński Ł J, Hagedorn PH, Lindow M, Vinther J (2017) RNase H sequence preferences influence antisense oligonucleotide efficiency. Nucleic Acids Res 45(22):12932–12944
Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA (2000) The sequential mechanism of HIV reverse transcriptase RNase H. J Biol Chem 275(48):37664–37671
Wang SX, Liu KS, Lou YF, Wang SQ, Peng YB, Chen JP, Huang JH, Xie SX, Cui L, Wang X (2018) RNase H meets molecular beacons: an ultrasensitive fluorometric assay for nucleic acids. Microchim Acta 185(8):375
Ma C, Wu K, Han Z, Liu H, Wang K, Xia K (2018) Fluorometric aptamer-based determination of ochratoxin A based on the use of graphene oxide and RNase H-aided amplification. Microchim Acta 185(7):347
Kanaya E, Kanaya S (1995) Kinetic analysis of Escherichia coli ribonuclease HI using oligomeric DNA/RNA substrates suggests an alternative mechanism for the interaction between the enzyme and the substrate. Eur J Biochem 231(3):557–562
Hogrefe HH, Hogrefe R, Walder R, Walder J (1990) Kinetic analysis of Escherichia coli RNase H using DNA-RNA-DNA/DNA substrates. J Biol Chem 265(10):5561–5566
Xie X, Xu W, Li T, Liu X (2011) Colorimetric detection of HIV-1 ribonuclease H activity by gold nanoparticles. Small 7(10):1393–1396
Chang YL, Kang KS, Park KS, Park HG (2018) Determination of RNase H activity via real-time monitoring of target-triggered rolling circle amplification. Microchim Acta 185(1):53
Lee CY, Jang H, Park KS, Park HG (2017) A label-free and enzyme-free signal amplification strategy for a sensitive RNase H activity assay. Nanoscale 9(42):16149–16153
Hu D, Pu F, Huang Z, Ren J, Qu X (2010) A Quadruplex-based, label-free, and real-time fluorescence assay for RNase H activity and inhibition. Chem Eur J 16(8):2605–2610
Wu K, Ma C, Liu H, He H, Zeng W, Wang K (2017) Label-free fluorescence assay for rapid detection of RNase H activity based on Tb 3+-induced G-quadruplex conjugates. Anal Methods 9(20):3055–3060
Si H, Sheng R, Li Q, Feng J, Li L, Tang B (2018) Highly sensitive fluorescence imaging of Zn2+ and Cu2+ in living cells with signal amplification based on functional DNA self-assembly. Anal Chem 90(15):8785–8792
McGhee CE, Loh KY, Lu Y (2017) DNAzyme sensors for detection of metal ions in the environment and imaging them in living cells. Curr Opin Biotechnol 45:191–201
Peng H, Newbigging AM, Wang Z, Tao J, Deng W, Le XC, Zhang H (2017) DNAzyme-mediated assays for amplified detection of nucleic acids and proteins. Anal Chem 90(1):190–207
Peng H, Li XF, Zhang H, Le XC (2017) A microRNA-initiated DNAzyme motor operating in living cells. Nat Commun 8:14378
Alizadeh N, Hallaj R, Salimi A (2017) A highly sensitive electrochemical immunosensor for hepatitis B virus surface antigen detection based on Hemin/G-quadruplex horseradish peroxidase-mimicking DNAzyme-signal amplification. Biosens Bioelectron 94:184–192
Chen F, Bai M, Cao K, Zhao Y, Cao X, Wei J, Wu N, Li J, Wang L, Fan C (2017) Programming enzyme-initiated autonomous DNAzyme Nanodevices in living cells. ACS Nano 11(12):11908–11914
Wang L, Zhou H, Liu B, Zhao C, Fan J, Wang W, Tong C (2017) Fluorescence assay for ribonuclease H based on nonlabeled substrate and DNAzyme assisted Cascade amplification. Anal Chem 89(20):11014–11020
Kim JH, Estabrook RA, Braun G, Lee BR, Reich NO (2007) Specific and sensitive detection of nucleic acids and RNases using gold nanoparticle–RNA–fluorescent dye conjugates. Chem Commun (42):4342–4344
Zhao C, Fan J, Peng L, Zhao L, Tong C, Wang W, Liu B (2016) An end-point method based on graphene oxide for RNase H analysis and inhibitors screening. Biosens Bioelectron 90:103
Shi Y, Huang WT, Luo HQ, Li NB (2011) A label-free DNA reduced graphene oxide-based fluorescent sensor for highly sensitive and selective detection of hemin. Chem Commun 47(16):4676–4678
Tong C, Zhao C, Liu B, Li B, Ai Z, Fan J, Wang W (2018) Sensitive detection of RNase a activity and collaborative drug screening based on rGO and fluorescence probe. Anal Chem 90(4):2655–2661
Hu J, Liu M-h, Li Y, Tang B, C-y Z (2018) Simultaneous sensitive detection of multiple DNA glycosylases from lung cancer cells at the single-molecule level. Chem Sci 9(3):712–720
Chen Y, Yang CJ, Wu Y, Conlon P, Kim Y, Lin H, Tan W (2008) Light-switching excimer Beacon assays for ribonuclease H kinetic study. ChemBioChem 9(3):355–359
Rizzo J, Gifford L, Zhang X, Gewirtz A, Lu P (2002) Chimeric RNA–DNA molecular beacon assay for ribonuclease H activity. Mol Cell Probes 16(4):277–283
Villa JA, Pike DP, Patel KB, Lomonosova E, Lu G, Abdulqader R, Tavis JE (2016) Purification and enzymatic characterization of the hepatitis B virus ribonuclease H, a new target for antiviral inhibitors. Antivir Res 132:186–195
Beilhartz GL, Götte M (2010) HIV-1 ribonuclease H: structure, catalytic mechanism and inhibitors. Viruses 2(4):900–926
Babu CS, Dudev T, Lim C (2013) Differential role of the protein matrix on the binding of a catalytic aspartate to Mg2+ vs Ca2+: application to ribonuclease H. J Am Chem Soc 135(17):6541–6548
Kanaya S, Kohara A, Miura Y, Sekiguchi A, Iwai S, Inoue H, Ohtsuka E, Ikehara M (1990) Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem 265(8):4615–4621
Wang L, Tang J, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z (2018) 6-Biphenylmethyl-3-hydroxypyrimidine-2, 4-diones potently and selectively inhibited HIV reverse transcriptase-associated RNase H. Eur J Med Chem 156:680–691
Acknowledgements
This work was partially supported by the Natural Science Foundation of Hunan Province (h14JJ2049), the Natural Science Foundation of China (81501218, 81673579, 81874369 and 31672457).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Human serum samples used in this study were collected from healthy volunteers and patients. This work was performed with the written informed consent of health and patients. The studies were approved by the medical Ethics Committee of People’s Hospital of Hunan Province and performed in accordance with the ethical standards.
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors wish it to be known that, in their opinion, Chunyi Tong and Ting Zhou should be regarded as joint First Authors.
Electronic supplementary material
ESM 1
(DOCX 3.56 mb)
Rights and permissions
About this article
Cite this article
Tong, C., Zhou, T., Zhao, C. et al. Fluorometric determination of RNase H via a DNAzyme conjugated to reduced graphene oxide, and its application to screening for inhibitors and activators. Microchim Acta 186, 335 (2019). https://doi.org/10.1007/s00604-019-3425-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3425-6