Abstract
A method is described for the determination of the activity of endonuclease. It based on the deaggregation of gold nanoparticles (AuNPs) aggregated by the action of poly(diallyldimethylammonium chloride) (PDDA). A single-stranded DNA (ssDNA) is released after enzymatic cleavage catalyzed by endonuclease. The released fragments bind electrostatically to PDDA and inhibit the PDDA-induced aggregation of AuNPs. This is accompanied by a color change from blue to red and a decrease in the absorption ratio (A630/A520). Under the optimal conditions, this ratio increases linearly in the 0.001 to 1 U·μL−1 EcoRI endonuclease activity range. The detection limit is of 2 × 10−4 U·μL−1 which is much better or at least comparable to previous reports. The method is deemed to have wide scope in that it may be used to study other endonuclease activity (such as BamHI) by simply changing the specific recognition site of the hairpin-like DNA probe. The assay may also be employed to screening for inhibitors of EcoRI endonuclease.

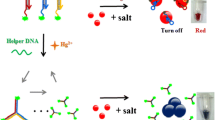
Schematic presentation of the colorimetric assay based on the deaggregation of AuNPs for the detection of endonuclease activity. A single-stranded sequence (ssDNA) is released by the EcoRI cleavage, which electrostatically binds to PDDA and inhibits the PDDA-induced aggregation of AuNPs accompanying with a color change from blue to red.





Similar content being viewed by others
References
Lieber MR (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays 19:233–240
Grindley ND, Whiteson KL, Rice PA (2006) Mechanisms of site-specific recombination. Annu Rev Biochem 75:567–605
Ma M, Benimetskaya L, Lebedeva I, Dignam J, Takle G, Stein CA (2000) Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat Biotechnol 18:58–61
Marti TM, Fleck O (2004) DNA repair nucleases. Cell Mol Life Sci 61:336–354
VanderVeen LA, Druckova A, Riggins JN, Sorrells JL, Guengerich FP, Marnett LJ (2005) Differential DNA recognition and cleavage by EcoRI dependent on the dynamic equilibrium between the two forms of the malondialdehyde-deoxyguanosine adduct. Biochemistry 44:5024–5033
Spitzer S, Eckstein F (1988) Inhibition of deoxyribonucleases by phosphorothioate groups in oligodeoxyribonucleotides. Nucleic Acids Res 16:11691–11704
Alves J, Rulter T, Geiger R, Fliess A, Maass G, Pingoud A (1989) Changing the hydrogen-bonding potential in the DNA binding site of EcoRI by site-directed mutagenesis drastically reduces the enzymic activity, not, however, the preference of this restriction endonuclease for cleavage within the site -GAATTC. Biochemistry 28:2678–2684
Li J, Yan H, Wang K, Tan W, Zhou X (2007) Hairpin fluorescence DNA probe for real-time monitoring of DNA methylation. Anal Chem 79:1050–1056
Li Y, LiY WY, Lu F, Chen Y, Gao W (2017) An electrochemiluminescence biosensor for endonuclease EcoRI detection. Biosens Bioelectron 89:585–591
Sun B, Jiang X, Wang H, Song B, Zhu Y, Wang H, Su Y, He Y (2015) Surface-enhancement Raman scattering sensing strategy for discriminatingtrace mercuric ion(II) from real water samples in sensitive specific, recyclable, and reproducible manners. Anal Chem 87:1250–1256
Sang F, Li X, Zhang Z, Liu J, Chen G (2018) Recyclable colorimetric sensor of Cr3+ and Pb2+ ions simultaneously using a zwitterionic amino acid modified gold nanoparticles. Spectrochim Part A 193:109–116
Chen L, Ye Y, Tan H, Wang Y (2015) A simple and rapid colorimetric method for the determination of Mn2+ based on pyrophosphate modified silver nanoparticles. Colloids Surf A Physicochem Eng Asp 478:1–6
He Y, Cheng F, Pang DW, Tang HW (2017) Colorimetric and visual determination of DNase I activity using gold nanoparticles as an indicator. Microchim Acta 184:1–6
Li T, Li Y, Zhang Y, Dong C, Shen Z, Wu A (2015) A colorimetric nitritedetection system with excellent selectivity and high sensitivity based on ag@au nanoparticles. Analyst 140:1076–1081
Sang F, Liu J, Zhang X, Pan J (2018) An aptamer-based colorimetric Pt(II) assay based on the use of gold nanoparticles and a cationic polymer. Microchim Acta 185:267
Kuang H, Chen W, Yan W, Xu L, Zhu Y, Liu L, Chu H, Peng C, Wang L, Kotov NA, Xu C (2011) Crown ether assembly of gold nanoparticles: melamine sensor. Biosens Bioelectron 26:2032–2037
Yao Y, Tian D, Li H (2010) Cooperative binding of bifunctionalized andclick-synthesized silver nanoparticles for colorimetric Co2+ sensing. ACS Appl Mater Interfaces 2:684–690
Kanaras A, Wang Z, Brust M, Cosstick R, Bates AD (2007) Enzymatic disassembly of DNA-gold nanostructures. Small 3:590–594
Song G, Chen C, Ren J, Qu X (2009) A simple, universal colorimetric assay for endonuclease/methyltransferase activity and inhibition based on an enzyme-responsive nanoparticle system. ACS Nano 3:1183–1189
Zhao W, Lam JCF, Chiuman W, Brook MA, Li Y (2008) Enzymatic cleavage of nucleic acids on gold nanoparticles: ageneric platform for facile colorimetric biosensors. Small 4:810–816
Liu Q, Li L, Zhao Y, Chen Z (2018) Colorimetric detection of DNA atthe nanomolar level based on enzyme-induced gold nanoparticle de-aggregation. Microchim Acta 185:301
Liu S, Du Z, Li P, Li F (2012) Sensitive colorimetric visualization of dihydronicotinamide adenine dinucleotide based on anti-aggregation of gold nanoparticles via boronic acid-diol binding. Biosens Bioelectron 35:443–446
Reyes M, Piotrowski M, Ang SK, Chan J, He S, Chu JJH, Kah JCY (2017) Exploiting the anti-aggregation of gold nanostars for rapid detection of hand, foot, and mouth disease causing enterovirus 71 using surface-enhanced raman spectroscopy. Anal Chem 89:5373–5381
Zhang J, Shi Z, Jin Y (2015) Enzyme-free and label-free signal amplification for monitoring endonuclease activity and inhibition via hybridization chain reaction. Analyst 140:3500–3506
Zheng J, Petty JT, Dickson RM (2003) High quantum yield blue emission from water-soluble Au8nanodots. J Am Chem Soc 125:7780–7781
Wang X, Gao W, Xu W, Xu S (2013) Fluorescent ag nanoclusters templated by carboxymethyl-β-cyclodextrin (CM-β-CD) and their in vitro antimicrobial activity. Mater Sci Eng C 33:656–662
Zhong Y, Deng C, He Y, Ge Y, Song G (2016) Exploring a monothiolated beta-cyclodextrin as the template to synthesize copper nanoclusters with exceptionally increased peroxidase-like activity. Microchim Acta 183:2823–2830
Jiang XQ, Guo SM, Zhang M, Zhou M, Ye BC (2014) DNA-hosted hoechst dyes: application for label-free fluorescent monitoring of endonucleaseactivity and inhibition. Analyst 139:5682–5685
Deng J, Jin Y, Chen G, Wang L (2012) Label-free fluorescent assay for real-time monitoring site-specific DNA cleavage by EcoRI endonuclease. Analyst 137:1713–1717
Deng J, Jin Y, Wang L, Chen G, Zhang C (2012) Sensitive detection of endonuclease activity and inhibition using gold nanorods. Biosens Bioelectron 34:144–150
Qian Y, Zhang Y, Lu L, Cai Y (2014) A label-free DNA-templated silver nanocluster probe for fluorescence on-off detection of endonuclease activity and inhibition. Biosens Bioelectron 51:408–412
Du YC, Zhu LN, Kong DM (2016) Label-free thioflavin T/G-quadruplex-based real-time strand displacement amplification for biosensing applications. Biosens Bioelectron 86:811–817
Acknowledgments
This work was funded by the Natural Science Foundation of China (NSFC) (No. 21407035), Shandong Provincial Natural Science Foundation (ZR2014BM021), Technology and Development Program of Weihai (2014DXGJ15), HIT-NSRIF (2011101).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures were in compliance with ethical strandards.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 2299 kb)
Rights and permissions
About this article
Cite this article
Sang, F., Li, G., Li, J. et al. A hairpin-type DNA probe for direct colorimetric detection of endonuclease activity and inhibition based on the deaggregation of gold nanoparticles. Microchim Acta 186, 100 (2019). https://doi.org/10.1007/s00604-018-3164-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-018-3164-0