Abstract
A photoelectrochemical (PEC) method is described for the determination of the activity of M.SssI methyltransferase (MTase). The assay relies on enzyme-linkage reactions and a DNA intercalator Ru(bpy)2(dppz)2+ (where bpy is 2,2′-bipyridine, and dppz is dipyrido[3,2-a:2′,3′-c]phenazine) which both serves as a PEC signal. The PEC electrode was obtained by immobilizing 5′-amino modified DNA strands (containing the methylation recognition site 5′-CCGG-3′) on a polyethylenimine (PEI) coated ITO/SnO2 electrode with glutaraldehyde as crosslinking agent. In the presence of MTase and S-adenosyl-L-methionine, the 5′-CCGG-3′ sequence in the DNA on the electrode is methylated. This protects the DNA strands from the shear of the methylation-sensitive restriction endonuclease HpaII. Consequently, more intact DNA strands remain on the surface of the electrode, providing more sites for Ru(bpy)2(dppz)2+ binding which in turn results in a high PEC response. The result demonstrates that the photocurrent increases linearly with the activity of MTase from 5 to 80 U·mL−1, and the limit of detection is 0.45 U·mL−1. The other MTases does not enhance the photocurrent, suggesting good selectivity of the assay. The method was also applied to rapid evaluate and screen the inhibitors of MTase. This strategy can be utilized to determinate the activity of other DNA MTases with specific DNA sequence.

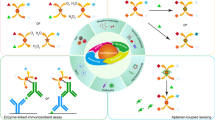
Schematic presentation of a photoelectrochemical assay based on enzyme-linkage reactions and a photo electrochemical probe combined with the oxalic acid involved cyclic amplification system for the determination of methyltransferase activity.






Similar content being viewed by others
References
Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S (2000) DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer 91:205–212
Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H, Mukai T (1991) A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci U S A 88:9523–9527
Xu Z, Yin H, Tian Z, Zhou Y, Ai S (2014) Electrochemical immunoassays for the detection the activity of DNA methyltransferase by using the rolling circle amplification technique. Microchim Acta 181:471–477
Wang H, Liu P, Jiang W, Li X, Yin H, Ai S (2017) Photoelectrochemical immunosensing platform for M.SssI methyltransferase activity analysis and inhibitor screening based on g-C3N4 and CdS quantum dots. Sensors Actuators B Chem 244:458–465
Miyamoto K, Fukutomi T, Akashi TS, Hasegawa T, Asahara T, Sugimura T (2005) Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer 116:407–414
Zhang Y, Wang X, Zhang Q, Zhang C (2017) Label-free sensitive detection of DNA methyltransferase by target-induced hyperbranched amplification with zero background signal. Anal Chem 89:12408–12415
Bergerat A, Guschlbauer W, Fazakerley GV (1991) Allosteric and catalytic binding of S-adenosylmethionine to Escherichia coli DNA adenine methyltransferase monitored by 3H NMR. Proc Natl Acad Sci U S A 88:6394–6397
Friso S, Choi S, Dolnikowski GG, Selhub J (2002) A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem 74:4526–4531
Zhao Y, Chen F, Wu Y, Dong Y, Fan C (2013) Highly sensitive fluorescence assay of DNA methyltransferase activity via methylation-sensitive cleavage coupled with nicking enzyme-assisted signalamplification. Biosens Bioelectron 42:56–61
Li W, Liu Z, Lin H, Nie Z, Chen J, Xu X (2010) Label-free colorimetric assay for methyltransferase activity based on a novel methylation-responsive dnazyme strategy. Anal Chem 82:1935–1941
Wu Z, Wu Z, Tang H, Tang L, Jiang J (2013) Activity-based dna-gold nanoparticle probe as colorimetric biosensor for dna methyltransferase/glycosylase assay. Anal Chem 85:4376–4383
Ouyang X, Liu J, Li J, Yang R (2012) A carbon nanoparticle-based low-background biosensing platform for sensitive and label-free fluorescent assay of DNA methylation. Chem Commun 48:88–90
Xu Z, Yin H, Tian Z, Zhou Y, Ai S (2014) Electrochemical immunoassays for the detection the activity of DNA methyltransferase by using the rolling circle amplification technique. Microchim Acta 181:471–477
Gao F, Fan T, Ou S, Wu J, Zhang X, Luo J, Li N, Yao Y, Mou Y, Liao X, Geng D (2018) Highly efficient electrochemical sensing platform for sensitive detection DNA methylation, and methyltransferase activity based on ag NPs decorated carbon nanocubes. Biosens Bioelectron 99:201–208
Shen Q, Han L, Fan G, Abdel-Halim E, Jiang L, Zhu J (2015) Highly sensitive photoelectrochemical assay for DNA methyltransferase activity and inhibitor screening by exciton energy transfer coupled with enzyme cleavage biosensing strategy. Biosens Bioelectron 64:449–455
Yang Z, Wang F, Wang M, Yin H, Ai S (2015) A novel signal-on strategy for M.SssI methyltransfease activity analysis and inhibitor screening based on photoelectrochemical immunosensor. Biosens Bioelectron 66:109–114
Cheng W, Pan J, Yang J, Zheng Z, Lu F, Chen Y (2018) A photoelectrochemical aptasensor for thrombin based on the use of carbon quantum dot-sensitized TiO2 and visible-light photoelectrochemical activity. Microchim Acta 185:263
Musumeci S, Rizzarelli F, Sammartano S, Bonomo R (1973) Low valence state of metal chelates. I. complexes of iron (II) perchlorate with 1, 10-phenanthroline, 4, 7-dimethyl-1, 10-phenanthroline and 4, 7-diphenyl-1,10-phenanthroline. Inorg. Chim. Acta 7:660–664
Zhang H, Dong H, Yang G, Chen H, Cai C (2016) Sensitive electrochemical detection of human methyltransferase based on a dual signal amplification strategy coupling gold nanoparticle-DNA complexes with Ru(III) redox recycling. Anal Chem 88:11108–11114
Liang M, Liu S, Wei M, Guo L (2006) Photoelectrochemical oxidation of DNA by ruthenium tris (bipyridine) on a tin oxide nanoparticle electrode. Anal Chem 78:621–623
Deng H, Yang X, Yeo S, Gao Z (2014) Highly sensitive electrochemical methyltransferase activity assay. Anal Chem 86:2117–2123
Liu Y, Liu Y, Xing Y, Guo X, Ye Y, Wu Y, Wen Y, Yang H (2018) Magnetically three-dimensional au nanoparticles/reduced graphene/ nickel foams for Raman trace detection. Sensors Actuators B Chem 273:884–890
Liu X, Xie X, Wei Y, Mao C, Chen J, Niu H (2018) Photoelectrochemical immunoassay for human interleukin 6 based on the use of perovskite-type LaFeO3 nanoparticles on fluorine-doped tin oxide glass. Microchim Acta 185:52
Wang Y, Bian F, Qin X, Wang Q (2018) Visible light photoelectrochemical aptasensor for chloramphenicol by using a TiO2 nanorod array sensitized with Eu(III)-doped CdS quantum dots. Microchim Acta 185:161
Zhang Z, Sheng S, Cao X, Li Y, Yao J, Wang T, Xie G (2015) Proximity-based electrochemical biosensor for highly sensitive determination of methyltransferase activity using gold nanoparticle-based cooperative signal amplification. Microchim Acta 182:2329–2336
Yang Z, Xie L, Yin H, Zhou Y, Ai S (2015) Methyltransferase activity assay based on the use of exonuclease III, the hemin/G-quadruplex system and reduced graphene oxide on a gold electrode, and a study on enzyme inhibition. Microchim Acta 182:2607–2613
Bi S, Zhao T, Luo B, Zhu J (2013) Hybridization chain reaction-based branched rolling circle amplification for chemiluminescence detection of DNA methylation. Chem Commun 49:6906–6908
Li X, Meng M, Zheng L, Xu Z, Song P, Yin Y (2016) Chemiluminescence immunoassay for s-adenosylhomocysteine detection and its application in dna methyltransferase activity evaluation and inhibitors screening. Anal Chem 88:8556–8561
Zhou H, Han T, Wei Q, Zhang S (2016) Efficient enhancement of electrochemiluminescence from cadmium sulfide quantum dots by glucose oxidase mimicking gold nanoparticles for highly sensitive assay of methyltransferase activity. Anal Chem 88:2976–2983
Acknowledgements
This work is supported by the National Natural Science Foundation of China (21475088, 21507087), PCSIRT (IRT1269), Chenguang Program of Shanghai Municipal Education Commission, and International Joint Laboratory on Resource Chemistry (IJLRC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
The supporting information is available free of charge. It includes the PEC response of the method with different concentrations of PEI; the PEC response of the method with different concentrations of glutaraldehyde; the photocurrent of Ru(bpy)2(dppz)2+ intercalated into the ITO/SnO2/PEI/GA/DNA film after it was incubated with different enzyme solution; the repeatability of the assay; the data of real sample detection
ESM 1
(DOCX 9255 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Wei, C., Luo, J. et al. Photoelectrochemical determination of the activity of M.SssI methyltransferase, and a method for inhibitor screening. Microchim Acta 185, 498 (2018). https://doi.org/10.1007/s00604-018-3033-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-018-3033-x