Abstract
The authors describe a method for DNA target recognition and signal amplification that is based on the target-induced formation of a three way junction. The subsequent assembly of two DNA probes releases the inhibitory strand and triggers a downstream strand displacement amplification. This causes the formation of a G-rich single sequence that binds to a hemin monomer with its peroxidase-mimicking properties. The resulting peroxidase (POx) activity is quantified by using H2O2 and TMB as the substrate. In the presence of an inhibitor, in contrast, the POx-like activity is strongly reduced. This forms the basis for a highly sensitive DNA assay. It has a 0.8 pM detection limit when operated at a wavelength of 450 nm and was applied to the isothermal determination of target DNA with high selectivity.

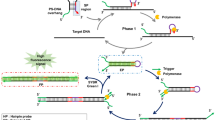
Schematic of the assay: Introduction of target results in the formation of a three way junction. The subsequent assembly of two probes releases the inhibitory strand and triggers a downstream strand displacement amplification, generating amount of G-rich single sequence which causes peroxidase-mimicking activity on binding to a hemin monomer.





Similar content being viewed by others
References
Wu J, Kodzius R, Cao W, Wen W (2014) Extraction, amplification and detection of DNA in microfluidic chip-based assays. Microchim Acta 181:1611–1631. doi:10.1007/s00604-013-1140-2
Khodakov D, Wang C, Zhang DY (2016) Diagnostics based on nucleic acid sequence variant profiling: PCR, hybridization, and NGS approaches. Adv Drug Deliv Rev. doi:10.1016/j.addr.2016.04.005
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction product by utilizing the 5'----3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A 88:7276–7280
Kevil CG, Walsh L, Laroux FS, Kalogeris T, Grisham MB, Alexander JS (1997) An improved, rapid northern protocol. Biochem Biophys Res Commun 238:277–279. doi:10.1006/bbrc.1997.7284
Volpi EV, Bridger JM (2008) FISH glossary: an overview of the fluorescence in situ hybridization technique. Biotechniques 45: 385–386, 388, 390. doi:10.2144/000112811
Bilitewski U (2009) DNA microarrays: an introduction to the technology. Methods Mol Biol 509:1–14. doi:10.1007/978-1-59745-372-1_1
Demidov VV, Frank-Kamenetskii MD (2004) Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem Sci 29:62–71. doi:10.1016/j.tibs.2003.12.007
Kolpashchikov DM (2010) Binary probes for nucleic acid analysis. Chem Rev 110:4709–4723. doi:10.1021/cr900323b
Marti AA, Jockusch S, Stevens N, Ju J, Turro NJ (2007) Fluorescent hybridization probes for sensitive and selective DNA and RNA detection. Acc Chem Res 40:402–409. doi:10.1021/ar600013q
Tang S, Tong P, Li H, Gu F, Zhang L (2013) The three-way junction DNAzyme based probe for label-free colorimetric detection of DNA. Biosens Bioelectron 41:397–402. doi:10.1016/j.bios.2012.08.056
Nakayama S, Yan L, Sintim HO (2008) Junction probes - sequence specific detection of nucleic acids via template enhanced hybridization processes. J Am Chem Soc 130:12560–12561. doi:10.1021/ja803146f
Zhang J, Chen JH, Chen RC, Chen GN, Fu FF (2009) Sequence-specific detection of trace DNA via a junction-probe electrochemical sensor employed template-enhanced hybridization strategy. Biosens Bioelectron 25:815–819. doi:10.1016/j.bios.2009.08.032
Wang Z, Zhang J, Guo Y, Wu X, Yang W, Xu L, Chen J, Fu F (2013) A novel electrically magnetic-controllable electrochemical biosensor for the ultra sensitive and specific detection of attomolar level oral cancer-related microRNA. Biosens Bioelectron 45:108–113. doi:10.1016/j.bios.2013.02.007
Li F, Lin Y, Le XC (2013) Binding-induced formation of DNA three-way junctions and its application to protein detection and DNA strand displacement. Anal Chem 85:10835–10841. doi:10.1021/ac402179a
Du Y, Jiang H, Huo Y, Han G, Kong D (2016) Optimization of strand displacement amplification-sensitized G-quadruplex DNAzyme-based sensing system and its application in activity detection of uracil-DNA glycosylase. Biosens Bioelectron 77:971–977. doi:10.1016/j.bios.2015.10.080
Zhang H, Liu Y, Gao J, Zhen J (2015) A sensitive SERS detection of miRNA using a label-free multifunctional probe. Chem Commun 51:16836–16839. doi:10.1039/C5CC06225J
Zhao Y, Zhou L, Tang Z (2013) Cleavage-based signal amplification of RNA. Nat Commun 4:1493. doi:10.1038/ncomms2492
Shi C, Liu Q, Ma C, Zhong W (2014) Exponential strand-displacement amplification for detection of MicroRNAs. Anal Chem 86:336–339. doi:10.1021/ac4038043
Li D, Cheng W, Yan Y, Zhang Y, Yin Y, Ju H, Ding S (2016) A colorimetric biosensor for detection of attomolar microRNA with a functional nucleic acid-based amplification machine. Talanta 146:470–476. doi:10.1016/j.talanta.2015.09.010
Ma C, Han D, Shi C (2014) A new isothermal nucleic acid detection strategy mediated by a double-nicked beacon. Chem Commun 50:3799. doi:10.1039/c3cc49841g
Cheng F, Jiang N, Li X, Zhang L, Hu L, Chen X, Jiang L, Abdel-Halim ES, Zhu J (2016) Target-triggered triple isothermal cascade amplification strategy for ultrasensitive microRNA-21 detection at sub-attomole level. Biosens Bioelectron 85:891–896. doi:10.1016/j.bios.2016.06.008
Zhang Q, Chen F, Xu F, Zhao Y, Fan C (2014) Target-triggered three-way junction structure and polymerase/nicking enzyme synergetic isothermal quadratic DNA machine for highly specific, one-step, and rapid MicroRNA detection at attomolar level. Anal Chem 86:8098–8105. doi:10.1021/ac501038r
Zhao Y, Qi L, Chen F, Zhao Y, Fan C (2013) Highly sensitive detection of telomerase activity in tumor cells by cascade isothermal signal amplification based on three-way junction and base-stacking hybridization. Biosens Bioelectron 41:764–770. doi:10.1016/j.bios.2012.10.009
Wang X, Liu W, Yin B, Yu P, Duan X, Liao Z, Liu C, Sang Y, Zhang G, Chen Y, Tao Z (2016) Colorimetric detection of gene transcript by target-induced three-way junction formation. Talanta 158:1–5. doi:10.1016/j.talanta.2016.05.039
Travascio P, Li Y, Sen D (1998) DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem Biol 5:505–517
Pelossof G, Tel-Vered R, Elbaz J, Willner I (2010) Amplified biosensing using the horseradish peroxidase-mimicking DNAzyme as an electrocatalyst. Anal Chem 82:4396–4402. doi:10.1021/ac100095u
Wang Q, Xu N, Gui Z, Lei J, Ju H, Yan F (2014) Catalytic activity of a dual-hemin labelled oligonucleotide: conformational dependence and fluorescent DNA sensing. Chem Commun 50:15362–15365. doi:10.1039/C4CC07298G
Grinberg LN, O'Brien PJ, Hrkal Z (1999) The effects of heme-binding proteins on the peroxidative and catalatic activities of hemin. Free Radic Biol Med 27:214–219
Miller YI, Felikman Y, Shaklai N (1995) The involvement of low-density lipoprotein in hemin transport potentiates peroxidative damage. Biochim Biophys Acta 1272:119–127
Rus’ OB, Puchkaev AV, Metelitsa DI (1996) Methemalbumin--a biocatalyst of aromatic amine oxidation by hydrogen peroxide. Biokhimiia 61:1813–1824
Marras SA, Kramer FR, Tyagi S (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal 14:151–156
Dubertret B, Calame M, Libchaber AJ (2001) Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat Biotechnol 19:365–370. doi:10.1038/86762
Mitelman F, Johansson B, Mertens F (2007) The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7:233–245. doi:10.1038/nrc2091
Edwards PA (2009) Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol n/a-n/a. doi:10.1002/path.2632
Lewis R (2004) Human genetics: concepts and applications, 6th edn. McGraw-Hill, Boston
Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA (2011) NUPACK: analysis and design of nucleic acid systems. J Comput Chem 32:170–173. doi:10.1002/jcc.21596
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant nos. 81271917), Natural Science Foundation of Zhejiang province (Grant nos. LY14H200002, LY15H200002 and LY16H160023). We thank Clinical Research Centre from the Second Affiliated Hospital of Zhejiang University School of Medicine for essential technical supports.
Author information
Authors and Affiliations
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Xuchu Wang and Weiwei Liu contributed equally to this work.
Electronic supplementary material
ESM 1
(DOC 237 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Liu, W., Yin, B. et al. An isothermal strand displacement amplification strategy for nucleic acids using junction forming probes and colorimetric detection. Microchim Acta 184, 1603–1610 (2017). https://doi.org/10.1007/s00604-017-2158-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2158-7