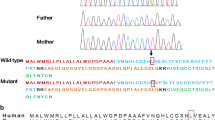
Abstract
Aim
To identify the genetic etiology of neonatal diabetes in an infant and to elucidate the molecular mechanism of the identified mutation underlying the pathogenesis.
Methods
Genetic analysis was carried out by sequencing of known etiological genes associated with NDM. Molecular characterization was performed by constructing a identified mutation in NKX2-2 gene and functional aspects was tested using transactivation, protein expression, DNA binding, nuclear localization assays. Structural analysis was performed by modeling the NKX2-2 protein structure.
Results
A novel homozygous frameshift mutation c.772delC, p.Q258SFs*59 in the NKX2-2 gene was identified in a patient with neonatal diabetes. Functional studies revealed that this mutation resulted in an elongated protein sequence, affecting DNA binding activity and transcriptional function. Structural analysis suggested alterations in the protein’s tertiary structure, likely contributing to its dysfunction.
Conclusion
This study presents the first report of a stop-loss mutation in the NKX2-2 gene associated with NDM. Our findings emphasize the importance of functional and structural characterization to understand the biological consequences of such mutations. This comprehensive analysis provides insights into the molecular mechanisms underlying NDM and its clinical phenotype, which may aid in better diagnosis and management of patients with similar variants in the future.


Similar content being viewed by others
References
De Franco E, Flanagan SE, Houghton JA, Lango Allen H, Mackay DJ, Temple IK, Ellard S, Hattersley AT (2015) The effect of early, comprehensive genomic testing on clinical care in neonatal diabetes: an international cohort study. Lancet (London, England) 386(9997):957–963. https://doi.org/10.1016/S0140-6736(15)60098-8
Iafusco D, Massa O, Pasquino B, Colombo C, Iughetti L, Bizzarri C, Mammì C, Lo Presti D, Suprani T, Schiaffini R, Nichols CG, Russo L, Grasso V, Meschi F, Bonfanti R, Brescianini S, Barbetti F, Early Diabetes Study Group of ISPED (2012) Minimal incidence of neonatal/infancy onset diabetes in Italy is 1:90,000 live births. Acta Diabetol 49(5):405–408. https://doi.org/10.1007/s00592-011-0331-8
Kanakatti Shankar R, Pihoker C, Dolan LM, Standiford D, Badaru A, Dabelea D, Rodriguez B, Black MH, Imperatore G, Hattersley A, Ellard S, Gilliam LK, SEARCH for Diabetes in Youth Study Group (2013) Permanent neonatal diabetes mellitus: prevalence and genetic diagnosis in the SEARCH for Diabetes in Youth Study. Pediatr Diabetes 14(3):174–180. https://doi.org/10.1111/pedi.12003
Greeley SAW, Polak M, Njølstad PR, Barbetti F, Williams R, Castano L, Raile K, Chi DV, Habeb A, Hattersley AT, Codner E (2022) ISPAD Clinical Practice Consensus Guidelines 2022: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes 23(8):1188–1211. https://doi.org/10.1111/pedi.13426
Ashcroft FM, Puljung MC, Vedovato N (2017) Neonatal diabetes and the KATP Channel: from mutation to therapy. Trends Endocrinol Metab 28(5):377–387. https://doi.org/10.1016/j.tem.2017.02.003
Pearson ER, Flechtner I, Njølstad PR, Malecki MT, Flanagan SE, Larkin B, Ashcroft FM, Klimes I, Codner E, Iotova V, Slingerland AS, Shield J, Robert JJ, Holst JJ, Clark PM, Ellard S, Søvik O, Polak M, Hattersley AT, Neonatal Diabetes International Collaborative Group (2006). Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. New Engl J Medi 355(5):467–477. https://doi.org/10.1056/NEJMoa061759
Gopi S, Kavitha B, Kanthimathi S, Kannan A, Kumar R, Joshi R, Kanodia S, Arya AD, Pendsey S, Pendsey S, Raghupathy P, Mohan V, Radha V (2021) Genotype-phenotype correlation of KATP channel gene defects causing permanent neonatal diabetes in Indian patients. Pediatr Diabetes 22(1):82–92. https://doi.org/10.1111/pedi.13109
Balamurugan K, Kavitha B, Yang Z, Mohan V, Radha V, Shyng SL (2019) Functional characterization of activating mutations in the sulfonylurea receptor 1 (ABCC8) causing neonatal diabetes mellitus in Asian Indian children. Pediatr Diabetes 20(4):397–407. https://doi.org/10.1111/pedi.12843
Flanagan SE, De Franco E, Lango Allen H, Zerah M, Abdul-Rasoul MM, Edge JA, Stewart H, Alamiri E, Hussain K, Wallis S, de Vries L, Rubio-Cabezas O, Houghton JA, Edghill EL, Patch AM, Ellard S, Hattersley AT (2014) Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab 19(1):146–154. https://doi.org/10.1016/j.cmet.2013.11.021
Auerbach A, Cohen A, Ofek Shlomai N, Weinberg-Shukron A, Gulsuner S, King MC, Hemi R, Levy-Lahad E, Abulibdeh A, Zangen D (2020) NKX2–2 mutation causes congenital diabetes and infantile obesity with paradoxical glucose-induced ghrelin secretion. J Clin Endocrinol Metabolism 105(11):dgaa563. https://doi.org/10.1210/clinem/dgaa563
Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS (1998) Mice lacking the homeodomain transcription factor Nkx2-2 have diabetes due to arrested differentiation of pancreatic beta cells. Development (Cambridge, England) 125(12):2213–2221. https://doi.org/10.1242/dev.125.12-2213
Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L (2004) Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci USA 101(9):2924–2929. https://doi.org/10.1073/pnas.0308604100
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Hwang KJ, Xiang B, Gruschus JM, Nam KY, No KT, Nirenberg M, Ferretti JA (2003) Distortion of the three-dimensional structure of the vnd/NK-2 homeodomain bound to DNA induced by an embryonically lethal A35T point mutation. Biochemistry 42(43):12522–12531. https://doi.org/10.1021/bi030116i
Gruschus JM, Tsao DH, Wang LH, Nirenberg M, Ferretti JA (1997) Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity. Biochemistry 36(18):5372–5380. https://doi.org/10.1021/bi9620060
Buchan DWA, Jones DT (2019) The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res 47(W1):W402–W407. https://doi.org/10.1093/nar/gkz297
Wilson ME, Scheel D, German MS (2003) Gene expression cascades in pancreatic development. Mech Dev 120(1):65–80
Doyle MJ, Sussel L (2007) Nkx2-2 regulates beta-cell function in the mature islet. Diabetes 56(8):1999–2007. https://doi.org/10.2337/db06-1766
Acknowledgements
Authors thank the patient and her parents for giving the blood samples for the study.
Funding
This study was supported by the Indian Council of Medical Research (ICMR), India, through the project Functional Studies on Variants of Pancreatic β-cell genes (HNF1A, HNF4A, ABCC8 and KCNJ11) in monogenic diabetes—an experimental approach with clinical translational potential; grant no: No. 5/4/5–2/Diab/2020-NCD-III awarded to VR.
Author information
Authors and Affiliations
Contributions
VR and BK conceptualized and implemented the study; BK performed and analyzed the functional data and wrote manuscript; NC and DS designed and performed structural analysis; NC analyzed the structural data; SG performed segregation analysis; KS and VM collected the clinical data and analyzed the manuscript; VR analyzed all data and corrected the manuscript. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from the proband parents included in the study.
Additional information
Managed by Fabrizio Barbetti.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kavitha, B., Srikanth, K., Singh, D. et al. A novel stop-loss mutation in NKX2-2 gene as a cause of neonatal diabetes mellitus: molecular characterization and structural analysis. Acta Diabetol 61, 189–194 (2024). https://doi.org/10.1007/s00592-023-02192-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-023-02192-y