Abstract
Aims
MicroRNA-103 (miR-103) family plays important roles in regulating glucose homeostasis in type 2 diabetes mellitus (DM2). However, the underlying mechanisms remain poorly characterized. The objective of this study was to test the hypothesis that circulating miR-103a and miR-103b, which regulate CAV-1 and SFRP4, respectively, are novel biomarkers for diagnosis of DM2.
Methods
We determined the predictive potential of circulating miR-103a and miR-103b in pre-DM subjects (pre-DM), noncomplicated diabetic subjects, and normal glucose-tolerance individuals (control) using bioinformatic analysis, qRT-PCR, luciferase assays, and ELISA assays.
Results
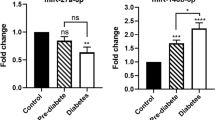
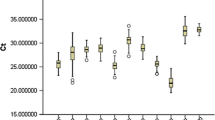
We found that both miR-103a and miR-103b had high complementarity and conservation, modulated reporter gene expression through seed sequences in the 3′UTRs of CAV-1 and SFRP4 mRNA, and negatively regulated their mRNA and protein levels, respectively. We also found that increased miR-103a and decreased miR-103a in plasma were significantly and negatively correlated with reduced CAV-1 levels and elevated SFRP4 levels in pre-DM and DM2, respectively, and were significantly associated with glucose metabolism, HbA1c levels, and other DM2 risk factors for progression from a normal individual to one with pre-DM. Furthermore, we demonstrated that the reciprocal changes in circulating miR-103a and miR-103b not only provided high sensitivity and specificity to differentiate the pre-DM population but also acted as biomarkers for predicting DM2 with high diagnostic value.
Conclusions
These findings suggest that circulating miR-103a and miR-103b may serve as novel biomarkers for diagnosis of DM2, providing novel insight into the mechanisms underlying pre-DM.








Similar content being viewed by others
Abbreviations
- DM2:
-
Type 2 diabetes mellitus
- miR-103:
-
MicroRNA-103
- NCDM:
-
Noncomplicated diabetic subjects
- pre-DM:
-
Pre-diabetes mellitus
- CAV-1:
-
Caveolin 1
- SFRP4:
-
Secreted frizzled-related protein 4
- FPG:
-
Fasting plasma glucose
- 2hPG:
-
2-hour post-load glucose
- OGTT:
-
Oral glucose-tolerance test
- BMI:
-
Body mass index
- WC:
-
Waist circumference
- TC:
-
Total cholesterol
- TG:
-
Triglyceride
- HDL-C:
-
High-density lipoprotein cholesterol
- LDL-C:
-
Low-density lipoprotein cholesterol
- SBP:
-
Systolic blood pressure
- DBP:
-
Diastolic blood pressure
- HbA1c:
-
Hemoglobin A1c
References
Chaudhury A, Duvoor C, Reddy Dendi VS, Kraleti S, Chada A, Ravilla R, Marco A, Shekhawat NS, Montales MT, Kuriakose K, Sasapu A, Beebe A, Patil N, Musham CK, Lohani GP, Mirza W (2017) Clinical review of antidiabetic drugs: implications for type 2 diabetes mellitus management. Front Endocrinol 8:6
Karalliedde J, Gnudi L (2016) Diabetes mellitus, a complex and heterogeneous disease, and the role of insulin resistance as a determinant of diabetic kidney disease. Nephrol Dial Transplant 31:206–213
American Diabetes A (2013) Diagnosis and classification of diabetes mellitus. Diabetes Care 36(Suppl 1):S67–S74
Bloomgarden ZT (2008) American College of Endocrinology pre-diabetes consensus conference: part three. Diabetes Care 31:2404–2409
Grundy SM (2012) Pre-diabetes, metabolic syndrome, and cardiovascular risk. J Am Coll Cardiol 59:635–643
Stepanek L, Horakova D, Nakladalova M, Cibickova L, Karasek D, Zadrazil J (2018) Significance of prediabetes as a nosological entity. Biomedical Papers of the Medical Faculty of the University Palacky, Olomouc
Tuso P (2014) Prediabetes and lifestyle modification: time to prevent a preventable disease. Perm J 18:88–93
Vaishya S, Sarwade RD, Seshadri V (2018) MicroRNA, proteins, and metabolites as novel biomarkers for prediabetes, diabetes, and related complications. Front Endocrinol 9:180
Guay C, Regazzi R (2013) Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 9:513–521
Sebastiani G, Nigi L, Grieco GE, Mancarella F, Ventriglia G, Dotta F (2017) Circulating microRNAs and diabetes mellitus: a novel tool for disease prediction, diagnosis, and staging? J Endocrinol Invest 40:591–610
Tiwari J, Gupta G, de Jesus Andreoli Pinto T, Sharma R, Pabreja K, Matta Y, Arora N, Mishra A, Sharma R, Dua K (2018) Role of microRNAs (miRNAs) in the pathophysiology of diabetes mellitus. Panminerva Med 60:25–28
Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y (2013) MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 33:178–185
Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, Shyy JY, Liang JT, Chen RH (2012) miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Can Res 72:3631–3641
Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M (2011) MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474:649–653
Wang JX, Zhang XJ, Li Q, Wang K, Wang Y, Jiao JQ, Feng C, Teng S, Zhou LY, Gong Y, Zhou ZX, Liu J, Wang JL, Li PF (2015) MicroRNA-103/107 regulate programmed necrosis and myocardial ischemia/reperfusion injury through targeting FADD. Circ Res 117:352–363
Woeller CF, Flores E, Pollock SJ, Phipps RP (2017) Editor’s highlight: Thy1 (CD90) expression is reduced by the environmental chemical tetrabromobisphenol-A to promote adipogenesis through induction of microRNA-103. Toxicol Sci 157:305–319
Wilfred BR, Wang WX, Nelson PT (2007) Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 91:209–217
Vatandoost N, Amini M, Iraj B, Momenzadeh S, Salehi R (2015) Dysregulated miR-103 and miR-143 expression in peripheral blood mononuclear cells from induced prediabetes and type 2 diabetes rats. Gene 572:95–100
Luo M, Li R, Deng X, Ren M, Chen N, Zeng M, Yan K, Xia J, Liu F, Ma W, Yang Y, Wan Q, Wu J (2015) Platelet-derived miR-103b as a novel biomarker for the early diagnosis of type 2 diabetes. Acta Diabetol 52:943–949
Zhu H, Leung SW (2015) Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia 58:900–911
Chakraborty C, Doss CG, Bandyopadhyay S, Agoramoorthy G (2014) Influence of miRNA in insulin signaling pathway and insulin resistance: micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip Rev RNA 5:697–712
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucl Acids Res 47:D155–D162
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucl Acids Res 37:W202–W208
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, Apweiler R (2009) QuickGO: a web-based tool for gene ontology searching. Bioinformatics 25:3045–3046
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, Chiew MY, Tai CS, Wei TY, Tsai TR, Huang HT, Wang CY, Wu HY, Ho SY, Chen PR, Chuang CH, Hsieh PJ, Wu YS, Chen WL, Li MJ, Wu YC, Huang XY, Ng FL, Buddhakosai W, Huang PC, Lan KC, Huang CY, Weng SL, Cheng YN, Liang C, Hsu WL, Huang HD (2018) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucl Acids Res 46:D296–D302
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44:839–847
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucl Acids Res 43:D146–D152
Wang B (2013) Base composition characteristics of mammalian miRNAs. J Nucl Acids 2013:951570
Hsueh WA, Orloski L, Wyne K (2010) Prediabetes: the importance of early identification and intervention. Postgrad Med 122:129–143
Bergman M (2013) Pathophysiology of prediabetes and treatment implications for the prevention of type 2 diabetes mellitus. Endocrine 43:504–513
Dorsey R, Songer T (2011) Lifestyle behaviors and physician advice for change among overweight and obese adults with prediabetes and diabetes in the United States, 2006. Prev Chronic Disease 8:A132
Eastwood SV, Tillin T, Sattar N, Forouhi NG, Hughes AD, Chaturvedi N (2015) Associations between prediabetes, by three different diagnostic criteria, and incident CVD differ in South Asians and Europeans. Diabetes Care 38:2325–2332
Bhatia P, Raina S, Chugh J, Sharma S (2015) miRNAs: early prognostic biomarkers for Type 2 diabetes mellitus? Biomark Med 9:1025–1040
Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramírez CM, Goedeke L, Rotllan N, Bonora E, Hughes AD, Santer P, Fernández-Hernando C, Tilg H, Willeit J, Kiechl S, Mayr M (2017) Circulating microrna-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 66:347–357
Yan S, Wang T, Huang S, Di Y, Huang Y, Liu X, Luo Z, Han W, An B (2016) Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol 53:693–702
Al-Muhtaresh HA, Al-Kafaji G (2018) Evaluation of two-diabetes related microRNAs suitability as earlier blood biomarkers for detecting prediabetes and type 2 diabetes mellitus. J Clin Med 7
Villard A, Marchand L, Thivolet C, Rome S (2015) Diagnostic value of cell-free circulating microRNAs for obesity and type 2 diabetes: a meta-analysis. J Mol Biomark Diagn 6
Wang S, Wang N, Zheng Y, Zhang J, Zhang F, Wang Z (2017) Caveolin-1: an oxidative stress-related target for cancer prevention. Oxid Med Cell Long 2017:7454031
Feng H, Guo W, Han J, Li XA (2013) Role of caveolin-1 and caveolae signaling in endotoxemia and sepsis. Life Sci 93:1–6
Cohen AW, Combs TP, Scherer PE, Lisanti MP (2003) Role of caveolin and caveolae in insulin signaling and diabetes. Am J Physiol Endocrinol Metab 285:E1151–E1160
Catalan V, Gomez-Ambrosi J, Rodriguez A, Silva C, Rotellar F, Gil MJ, Cienfuegos JA, Salvador J, Frühbeck G (2008) Expression of caveolin-1 in human adipose tissue is upregulated in obesity and obesity-associated type 2 diabetes mellitus and related to inflammation. Clin Endocrinol 68:213–219
Carrillo-Sepulveda MA, Matsumoto T (2014) Phenotypic modulation of mesenteric vascular smooth muscle cells from type 2 diabetic rats is associated with decreased caveolin-1 expression. Cell Physiol Biochem 34:1497–1506
Briand N, Le Lay S, Sessa WC, Ferre P, Dugail I (2011) Distinct roles of endothelial and adipocyte caveolin-1 in macrophage infiltration and adipose tissue metabolic activity. Diabetes 60:448–453
Mahdi T, Hanzelmann S, Salehi A, Muhammed SJ, Reinbothe TM, Tang Y, Axelsson AS, Zhou Y, Jing X, Almgren P, Krus U, Taneera J, Blom AM, Lyssenko V, Esguerra JL, Hansson O, Eliasson L, Derry J, Zhang E, Wollheim CB, Groop L, Renström E, Rosengren AH (2012) Secreted frizzled-related protein 4 reduces insulin secretion and is overexpressed in type 2 diabetes. Cell Metab 16:625–633
Anand K, Vidyasagar S, Lasrado I, Pandey GK, Amutha A, Ranjani H, Mohan Anjana R, Mohan V, Gokulakrishnan K (2016) Secreted frizzled-related protein 4 (SFRP4): a novel biomarker of beta-cell dysfunction and insulin resistance in individuals with prediabetes and type 2 diabetes. Diabetes Care 39:e147–e148
Zou D, Ye Y, Zou N, Yu J (2017) Analysis of risk factors and their interactions in type 2 diabetes mellitus: a cross-sectional survey in Guilin, China. J Diabetes Investig 8:188–194
Liu L, Guan X, Yuan Z, Zhao M, Li Q, Zhang X, Zhang H, Zheng D, Xu J, Gao L, Guan Q, Zhao J, The Reaction Study Group (2019) Different contributions of dyslipidemia and obesity to the natural history of type 2 diabetes: 3-year cohort study in China. J Diabetes Res 2019:4328975
Biesenbach G (1989) Disorders of lipid metabolism in diabetes mellitus. Wien Med Wochenschr Suppl 105:9–17
Yang Z, Chen H, Si H, Li X, Ding X, Sheng Q, Chen P, Zhang H (2014) Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol 51:823–831
Al-Kafaji G, Al-Mahroos G, Alsayed NA, Hasan ZA, Nawaz S, Bakhiet M (2015) Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol Med Rep 12:7485–7490
Liu Y, Gao G, Yang C, Zhou K, Shen B, Liang H, Jiang X (2014) The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int J Mol Sci 15:10567–10577
Párrizas M, Brugnara L, Esteban Y, González-Franquesa A, Canivell S, Murillo S, Gordillo-Bastidas E, Cussó R, Cadefau JA, García-Roves PM, Servitja JM, Novials A (2015) Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J Clin Endocrinol Metab 100:E407–E415
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81800434, 81570263), Grant of Sichuan Province Science and Technology Agency Grant (2019YJ0487), Foundation of Luzhou Municipal Science and Technology Bureau (2017LZXNYD-T05, 2016LZXNYD-J24), and the Ministry Science and Technology of China Grant (2016YFC0901200, 2016YFC0901205).
Author information
Authors and Affiliations
Contributions
The original idea of this study was proposed by QW. ML and CX performed experiments and analyzed the data and wrote the first draft of this manuscript. YL and GW performed and interpreted the experiments. JW and QW edited subsequent drafts. All authors have read and approved the final version of the manuscript for submission.
Corresponding author
Ethics declarations
Conflict of interest
All the authors including Mao Luo, Chunrong Xu, Yulin Luo, Gang Wang, Jianbo Wu, and Qin Wan declare that they have no conflict of interest.
Ethical approval
All human subjects used in the study ‘‘Circulating miR-103 Family as Potential Biomarkers for Type 2 Diabetes through targeting CAV-1 and SFRP4’’ have been reviewed by the Research Ethics Committee of the Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan Province, P. R. China and have been performed in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All samples were collected with informed consent of all subjects. There is no security and privacy violation to the patient’s health in our study.
Human and animal rights
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the 1975 Helsinki declaration, as revised in 2008 (5).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Managed by Massimo Porta.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, M., Xu, C., Luo, Y. et al. Circulating miR-103 family as potential biomarkers for type 2 diabetes through targeting CAV-1 and SFRP4. Acta Diabetol 57, 309–322 (2020). https://doi.org/10.1007/s00592-019-01430-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-019-01430-6