Abstract
Aims
MicroRNAs (miRNAs) are present in plasma and have emerged as critical regulators of gene expression at posttranscriptional level, and thus are involved in various human diseases, including diabetes. The objective of this study was to screen and validate differentially expressed plasma miRNAs in prediabetes and newly diagnosed type 2 diabetes (T2D).
Methods
In this study, we screened differentially expressed plasma miRNAs in prediabetes and newly diagnosed T2D by miRNA microarray analysis, and validated the expression of candidate miRNAs using quantitative reverse transcription polymerase chain reaction assays. Furthermore, we performed gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses to disclose functional enrichment of genes predicted to be regulated by the differentially expressed miRNAs.
Results
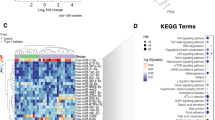
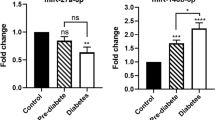
Notably, our results revealed that hsa-miR-1249, hsa-miR-320b, and hsa-miR-572 (P < 0.05) were differentially expressed among the three groups, which yielded an area under the receiver operator characteristics curve (AUC) of 0.784 [95 % confidence interval (CI) 0.685–0.883], 0.946 (95 % CI 0.906–0.985), and 0.843 (95 % CI 0.766–0.920) discriminating T2D patients from NGT control groups, respectively, while the AUC was 0.887 (95 % CI 0.818–0.957), 0.635 (95 % CI 0.525–0.744), and 0.69 (95 % CI 0.580–0.793) discriminating prediabetes patients from NGT control groups, respectively. In addition, GO and KEGG pathway analyses showed that genes predicted to be regulated by differentially expressed miRNAs were significantly enriched in several related biological processes and pathways, including the development of multicellular organisms, signal transduction, cell differentiation, apoptosis, cell metabolism, ion transport regulation, and other biological functions.
Conclusions
Taken together, our results showed differentially expressed miRNAs in T2D and prediabetes. Plasma hsa-miR-1249, hsa-miR-320b, and hsa-miR-572 may serve as novel biomarkers for diagnosis and potential targets for the treatment for prediabetes and T2D.





Similar content being viewed by others
References
ADA (American Diabetes Association) (2013) Diagnosis and classification of diabetes mellitus. Diabetes Care 36(Suppl 1):S67–S74
ADA (American Diabetes Association) (2013) Standards of medical care in diabetes—2013. Diabetes Care 36(Suppl 1):S11–S66
Afghahi H, Miftaraj M, Svensson AM et al (2013) Ongoing treatment with renin-angiotensin-aldosterone-blocking agents does not predict normoalbuminuric renal impairment in a general type 2 diabetes population. J Diabetes Complicat 27:229–234
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bentwich I, Avniel A, Karov Y et al (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37:766–770
Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120:21–24
Chen X, Ba Y, Ma L et al (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18:997–1006
Curaba J, Singh MB, Bhalla PL (2014) miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 65:1425–1438
Daimiel-Ruiz L, Klett-Mingo M, Konstantinidou V et al (2015) Dietary lipids modulate the expression of miR-107, an miRNA that regulates the circadian system. Mol Nutr Food Res 59:552–565
El OA, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E (2008) miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 57:2708–2717
Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE (2014) Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract 103:137–149
Guay C, Regazzi R (2015) Role of islet microRNAs in diabetes: which model for which question? Diabetologia 58:456–463
Haug BH, Henriksen JR, Buechner J et al (2011) MYCN-regulated miRNA-92 inhibits secretion of the tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma. Carcinogenesis 32:1005–1012
He C (2013) The establishment of fingerprint spectrum of circulating microRNAs in early stage diabetic nephropathy. Chongqing Medical University, Chongqing, pp 1–53
Igoillo-Esteve M, Marselli L, Cunha DA et al (2010) Palmitate induces a pro-inflammatory response in human pancreatic islets that mimics CCL2 expression by beta cells in type 2 diabetes. Diabetologia 53:1395–1405
Jacobs ME, Jeffers LA, Welch AK, Wingo CS, Cain BD (2013) The effect of aldosterone on the miRNA content of a murine inner medullary collecting duct cell. FASEB J 27:778
Kong L, Zhu J, Han W et al (2011) Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 48:61–69
Kong L, Zhu J, Han W et al (2014) Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol 51:823–831
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
Li X (2014) MiR-375, a microRNA related to diabetes. Gene 533:1–4
Li Y, Jiang Z, Xu L, Yao H, Guo J, Ding X (2011) Stability analysis of liver cancer-related microRNAs. Acta Biochim Biophys Sin (Shanghai) 43:69–78
Liu AM, Wang W, Luk JM (2014) miRNAs: new tools for molecular classification, diagnosis and prognosis of hepatocellular carcinoma. Hepatic Oncol 1:323–329
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Luo M, Li R, Deng X et al (2015) Platelet-derived miR-103b as a novel biomarker for the early diagnosis of type 2 diabetes. Acta Diabetol 52:943–949
Maharaj PD, Widen SG, Huang J, Wood TG, Thangamani S (2015) Discovery of mosquito saliva microRNAs during CHIKV infection. PLoS Negl Trop Dis 9:e3386
Nilsen TW (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 23:243–249
Papadopoulos T, Belliere J, Bascands JL, Neau E, Klein J, Schanstra JP (2015) miRNAs in urine: a mirror image of kidney disease? Expert Rev Mol Diagn 15:361–374
Raciti GA, Longo M, Parrillo L et al (2015) Understanding type 2 diabetes: from genetics to epigenetics. Acta Diabetol 52:821–827
Redova M, Sana J, Slaby O (2013) Circulating miRNAs as new blood-based biomarkers for solid cancers. Future Oncol 9:387–402
Salunkhe VA, Esguerra JL, Ofori JK et al (2015) Modulation of microRNA-375 expression alters voltage-gated Na(+) channel properties and exocytosis in insulin-secreting cells. Acta Physiol (Oxford) 213:882–892
Sebastiani G, Po A, Miele E et al (2015) MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol 52:523–530
Shi P (2013) Analysis of the expression of circulating microRNA spectral in diabetic nephropathy patients. China Medical University, Shenyang, pp 1–38
Tang X, Muniappan L, Tang G, Ozcan S (2009) Identification of glucose-regulated miRNAs from pancreatic beta cells reveals a role for miR-30d in insulin transcription. RNA 15:287–293
Tang Y, Banan A, Forsyth CB et al (2008) Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res 32:355–364
Tay HL, Plank M, Collison A, Mattes J, Kumar RK, Foster PS (2014) MicroRNA: potential biomarkers and therapeutic targets for allergic asthma? Ann Med 46:633–639
Whiting DR, Guariguata L, Weil C, Shaw J (2011) IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 94:311–321
Xie X, Lu J, Kulbokas EJ et al (2005) Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature 434:338–345
Yang W, Lu J, Weng J et al (2010) Prevalence of diabetes among men and women in China. N Engl J Med 362:1090–1101
Zampetaki A, Kiechl S, Drozdov I et al (2010) Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 107:810–817
Zhang GJ (2011) Silicon nanowire biosensor for ultrasensitive and label-free direct detection of miRNAs. Methods Mol Biol 676:111–121
Acknowledgments
This work was supported by Science and Technology Support Program of Guizhou Province (SY20123115 of Guizhou Branch). We thank all of the patients for agreeing to participate in our study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflicts of interest were declared.
Ethical standard
All human subjects used in the study have been reviewed by the Research Ethics Committee (People’s Hospital of Guizhou Province, Guiyang, Guizhou Province, P. R. China) and have therefore been performed in accordance with the ethical standards.
Human and animal rights disclosure
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Declaration of Helsinki 1975, as revised in 2008 (5).
Informed consent disclosure
Informed consent was obtained from all patients for being included in the study.
Additional information
Managed by Massimo Federici.
Shaoying Yan and Tianqiong Wang have contributed equally to the project.
Rights and permissions
About this article
Cite this article
Yan, S., Wang, T., Huang, S. et al. Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol 53, 693–702 (2016). https://doi.org/10.1007/s00592-016-0837-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-016-0837-1