Abstract
Purpose
The study evaluated the diagnostic performance of metagenomic next-generation sequencing (mNGS) as a diagnostic test for biopsy samples from patients with suspected spinal infection (SI) and compared the diagnostic performance of mNGS with that of microbial culture.
Methods
All patients diagnosed with clinical suspicion of SI were enrolled, and data were collected through a retrospective chart review of patient records. Biopsy specimens obtained from each patient were tested via mNGS and microbial culture. Samples were enriched for microbial DNA using the universal DNA extraction kit, whole-genome amplified, and sequenced using MGISEQ-200 instrument. After Low-quality reads removed, the remaining sequences for microbial content were analyzed and aligned using SNAP and kraken2 tools.
Results
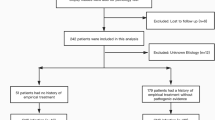
A total of 39 patients (19 men and 20 women) were deemed suitable for enrollment. The detection rate for pathogens of mNGS was 71.8% (28/39), which was significantly higher than that of microbial culture (23.1%, p = 0.016). Mycobacterium tuberculosis complex was the most frequently isolated. Using pathologic test as the standard reference for SI, thirty-one cases were classified as infected, and eight cases were considered aseptic. The sensitivity and specificity values for detecting pathogens with mNGS were 87.1% and 87.5%, while these rates were 25.8% and 87.5% with conventional culture. mNGS was able to detect 88.9% (8/9) of pathogens identified by conventional culture, with a genus-level sensitivity of 100% (8/8) and a species-level sensitivity of 87.5% (7/8).
Conclusion
The present work suggests that mNGS might be superior to microbial culture for detecting SI pathogens.



Similar content being viewed by others
References
Lener S, Wipplinger C, Hartmann S, Rietzler A, Thomé C, Tschugg A (2021) Gender-specific differences in presentation and management of spinal infection: a single-center retrospective study of 159 cases. Glob Spine J 11(4):430–436. https://doi.org/10.1177/2192568220905804
Nagashima H, Nanjo Y, Tanida A, Dokai T, Teshima R (2012) Clinical features of spinal infection in individuals older than eighty years. Int Orthop 36(6):1229–1234. https://doi.org/10.1007/s00264-011-1440-2
Kaya S, Ercan S, Kaya S et al (2014) Spondylodiscitis: evaluation of patients in a tertiary hospital. J Infect Dev Ctries 8(10):1272–1276. https://doi.org/10.3855/jidc.4522
Yeom JA, Lee IS, Suh HB, Song YS, Song JW (2016) Magnetic resonance imaging findings of early spondylodiscitis: interpretive challenges and atypical findings. Korean J Radiol 17(5):565–580. https://doi.org/10.3348/kjr.2016.17.5.565
Brown ED, Wright GD (2016) Antibacterial drug discovery in the resistance era. Nature 529(7586):336–343. https://doi.org/10.1038/nature17042
Liu J, Gefen O, Ronin I, Bar-Meir M, Balaban NQ (2020) Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367(6474):200–204. https://doi.org/10.1126/science.aay3041
Lopez J, Tournadre A, Couderc M, Pereira B, Soubrier M, Dubost JJ (2018) No change in the efficacy of infectious spondylodiscitis bacteriological testing over 20 years period. Joint Bone Spine 85(5):637–638. https://doi.org/10.1016/j.jbspin.2017.12.011
Ang MT, Wong GR, Wong DR, Clements W, Joseph T (2019) Diagnostic yield of computed tomography-guided biopsy and aspiration for vertebral osteomyelitis. J Med Imaging Radiat Oncol 63(5):589–595. https://doi.org/10.1111/1754-9485.12923
Czuczman GJ, Marrero DE, Huang AJ, Mandell JC, Ghazikhanian V, Simeone FJ (2018) Diagnostic yield of repeat CT-guided biopsy for suspected infectious spondylodiscitis. Skelet Radiol 47(10):1403–1410. https://doi.org/10.1007/s00256-018-2972-y
Lefterova MI, Suarez CJ, Banaei N, Pinsky BA (2015) Next-generation sequencing for infectious disease diagnosis and management: a report of the association for molecular pathology. J Mol Diagn 17(6):623–634. https://doi.org/10.1016/j.jmoldx.2015.07.004
Albanese D, Donati C (2017) Strain profiling and epidemiology of bacterial species from metagenomic sequencing. Nat Commun 8(1):2260. https://doi.org/10.1038/s41467-017-02209-5
Waseem H, Williams MR, Stedtfeld T et al (2017) Virulence factor activity relationships (VFARs): a bioinformatics perspective. Environ Sci Process Impacts 19(3):247–260. https://doi.org/10.1039/c6em00689b
Gu W, Miller S, Chiu CY (2019) Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 14:319–338. https://doi.org/10.1146/annurev-pathmechdis-012418-012751
Govender KN, Street TL, Sanderson ND, Eyre DW (2021) Metagenomic sequencing as a pathogen-agnostic clinical diagnostic tool for infectious diseases: a systematic review and meta-analysis of diagnostic test accuracy studies. J Clin Microbiol 59(9):e0291620. https://doi.org/10.1128/JCM.02916-20
Tang Y, Zhao D, Wang S, Yi Q, Xia Y, Geng B (2022) Diagnostic value of next-generation sequencing in periprosthetic joint infection: a systematic review. Orthop Surg 14(2):190–198. https://doi.org/10.1111/os.13191
Abboud Z, Galuppo L, Tolone M et al (2021) Molecular characterization of antimicrobial resistance and virulence genes of bacterial pathogens from bovine and caprine mastitis in northern lebanon. Microorganisms 9(6):1148. https://doi.org/10.3390/microorganisms9061148
Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE et al (2018) Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin Infect Dis 67(9):1333–1338. https://doi.org/10.1093/cid/ciy303
Pereira MB, Wallroth M, Jonsson V, Kristiansson E (2018) Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genom 19(1):274. https://doi.org/10.1186/s12864-018-4637-6
Ma C, Wu H, Chen G, Liang C, Wu L, Xiao Y (2022) The potential of metagenomic next-generation sequencing in diagnosis of spinal infection: a retrospective study. Eur Spine J 31(2):442–447. https://doi.org/10.1007/s00586-021-07026-5
Garg V, Kosmas C, Young PC, Togaru UK, Robbin MR (2014) Computed tomography-guided percutaneous biopsy for vertebral osteomyelitis: a department’s experience. Neurosurg Focus 37(2):E10. https://doi.org/10.3171/2014.6.FOCUS14134
Kim BJ, Lee JW, Kim SJ, Lee GY, Kang HS (2013) Diagnostic yield of fluoroscopy-guided biopsy for infectious spondylitis. AJNR Am J Neuroradiol 34(1):233–238. https://doi.org/10.3174/ajnr.A3120
Duarte RM, Vaccaro AR (2013) Spinal infection: state of the art and management algorithm. Eur Spine J 22(12):2787–2799. https://doi.org/10.1007/s00586-013-2850-1
McNamara AL, Dickerson EC, Gomez-Hassan DM, Cinti SK, Srinivasan A (2017) Yield of image-guided needle biopsy for infectious discitis: a systematic review and meta-analysis. AJNR Am J Neuroradiol 38(10):2021–2027. https://doi.org/10.3174/ajnr.A5337
Frey KG, Herrera-Galeano JE, Redden CL et al (2014) Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood. BMC Genom 15:96. https://doi.org/10.1186/1471-2164-15-96
Wang K, Liu X, Liu H et al (2021) Metagenomic diagnosis of severe psittacosis using multiple sequencing platforms. BMC Genom 22(1):406. https://doi.org/10.1186/s12864-021-07725-9
Thoendel M, Jeraldo P, Greenwood-Quaintance KE et al (2020) Comparison of three commercial tools for metagenomic shotgun sequencing analysis. J Clin Microbiol 58(3):e00981-e1019. https://doi.org/10.1128/JCM.00981-19
Agarwal V, Wo S, Lagemann GM, Tsay J, Delfyett WT (2016) Image-guided percutaneous disc sampling: impact of antecedent antibiotics on yield. Clin Radiol 71(3):228–234. https://doi.org/10.1016/j.crad.2015.10.031
Lüftinger L, Ferreira I, Frank B et al (2021) Predictive antibiotic susceptibility testing by next-generation sequencing for periprosthetic joint infections: potential and limitations. Biomedicines 9(8):910. https://doi.org/10.3390/biomedicines9080910
NIHR Global Health Research Unit on Genomic Surveillance of AMR (2020) Whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: a roadmap. BMJ Glob Health 5(11):e002244. https://doi.org/10.1136/bmjgh-2019-002244
Hendriksen RS, Bortolaia V, Tate H, Tyson GH, Aarestrup FM, McDermott PF (2019) Using genomics to track global antimicrobial resistance. Front Public Health 7:242. https://doi.org/10.3389/fpubh.2019.00242
Leal NC, Campos TL, Rezende AM et al (2020) Comparative genomics of Acinetobacter baumannii clinical strains from Brazil reveals polyclonal dissemination and selective exchange of mobile genetic elements associated with resistance genes. Front Microbiol 11:1176. https://doi.org/10.3389/fmicb.2020.01176
Hamidian M, Nigro SJ, Hall RM (2012) Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother 67(12):2833–2836. https://doi.org/10.1093/jac/dks318
Street TL, Sanderson ND, Atkins BL et al (2017) Molecular diagnosis of orthopedic-device-related infection directly from sonication fluid by metagenomic sequencing. J Clin Microbiol 55(8):2334–2347. https://doi.org/10.1128/JCM.00462-17
Kildow BJ, Ryan SP, Danilkowicz R et al (2021) Next-generation sequencing not superior to culture in periprosthetic joint infection diagnosis. Bone Joint J 103-B(1):26–31. https://doi.org/10.1302/0301-620X.103B1.BJJ-2020-0017.R3
Guo Z, Xiao D, Wang X, Wang Y, Yan T (2019) Epidemiological characteristics of pulmonary tuberculosis in mainland China from 2004 to 2015: a model-based analysis. BMC Public Health 19(1):219. https://doi.org/10.1186/s12889-019-6544-4
Kang W, Liu S, Du J et al (2022) Epidemiology of concurrent extrapulmonary tuberculosis in inpatients with extrapulmonary tuberculosis lesions in China: a large-scale observational multi-centre investigation. Int J Infect Dis 115:79–85. https://doi.org/10.1016/j.ijid.2021.11.019
Lecuit M, Eloit M (2015) The potential of whole genome NGS for infectious disease diagnosis. Expert Rev Mol Diagn 15(12):1517–1519. https://doi.org/10.1586/14737159.2015.1111140
Acknowledgements
We thank the patients and the patients’ guardians and families for their support. Without their participation, this report could not have been possible; we are very thankful. The mNGS assay was performed by a commercial institution (Guangzhou Daan Clinical Laboratory Center, China).
Funding
One author (ZW) has received funding from Guangdong Basic and Applied Basic Research Foundation (2021A1515011508) and the Scientific Research Start Plan of Shunde Hospital, Southern Medical University (CRSP2019010). Another author (WL) has received funding from the Scientific Research Start Plan of Shunde Hospital, Southern Medical University (SRSP2021007).
Author information
Authors and Affiliations
Contributions
WL: data curation; funding acquisition; investigation; writing—original draft; FX: data curation; supervision; validation; XL: writing—original draft; RY: data curation; investigation; methodology; project administration; JL: supervision; validation; writing—original draft; ZR: investigation; software; supervision; DO: formal analysis; investigation; methodology; software; ZW: conceptualization; funding acquisition; methodology; project administration; writing—review and editing.
Corresponding author
Ethics declarations
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Shunde Hospital, Southern Medical University (LWLS202201014).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, W., Xie, F., Li, X. et al. Diagnostic performance of metagenomic next-generation sequencing and conventional microbial culture for spinal infection: a retrospective comparative study. Eur Spine J 32, 4238–4245 (2023). https://doi.org/10.1007/s00586-023-07928-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-023-07928-6