Abstract
Purpose
To analyze the differential transcriptome expression in hypertrophic ligaments flavum (HLF) compared to normal ligaments.
Methods
A case–control study was conducted that included 15 patients with hypertrophy of LF and 15 controls. Samples of LF were obtained through a lumbar laminectomy and analyzed by DNA microarrays and histology. The dysregulated biological processes, signaling pathways, and pathological markers in the HLF were identified using bioinformatics tools.
Results
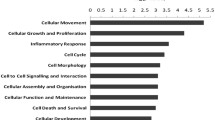
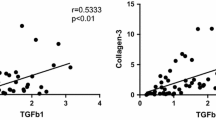
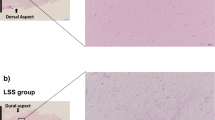
The HLF had notable histological alterations, including hyalinosis, leukocyte infiltration, and disarrangement of collagen fibers. Transcriptomic analysis showed that up-regulated genes were associated with the signaling pathways of Rho GTPases, receptor tyrosine kinases (RTK), fibroblast growth factors (FGF), WNT, vascular endothelial growth factor, phosphoinositide 3-kinase (PIK3), mitogen-activated protein kinases, and immune system. The genes PIK3R1, RHOA, RPS27A, CDC42, VAV1, and FGF5, 9, 18, and 19 were highlighted as crucial markers in HLF. The down-expressed genes in the HLF had associations with the metabolism of RNA and proteins.
Conclusion
Our results suggest that abnormal processes in hypertrophied LF are mediated by the interaction of the Rho GTPase, RTK, and PI3K pathways, which have not been previously described in the HLF, but for which there are currently therapeutic proposals. More studies are required to confirm the therapeutic potential of the pathways and mediators described in our results.



Similar content being viewed by others
References
Lee BH, Moon S-H, Suk K-S et al (2020) Lumbar spinal stenosis: pathophysiology and treatment principle: a narrative review. Asian Spine J 14:682–693. https://doi.org/10.31616/asj.2020.0472
Yoshiiwa T, Miyazaki M, Kawano M et al (2016) Analysis of the relationship between hypertrophy of the ligamentum flavum and lumbar segmental motion with aging process. Asian Spine J. https://doi.org/10.4184/asj.2016.10.3.528
Lee SY, Kim TH, Oh JK et al (2015) Lumbar stenosis: a recent update by review of literature. Asian Spine J 9(5):818–828
Williams MG, Wafai AM, Podmore MD (2017) Functional outcomes of laminectomy and laminotomy for the surgical management lumbar spine stenosis. J Spine Surg. https://doi.org/10.21037/jss.2017.10.08
Degen T, Fischer K, Theiler R et al (2020) Outcomes after spinal stenosis surgery by type of surgery in adults aged 60 years and older. Swiss Med Wkly. https://doi.org/10.4414/smw.2020.20325
Sidon E, Shemesh SS, Mor-Yossef Moldovan L et al (2019) Molecular profile of ultrastructure changes of the ligamentum flavum related to lumbar spinal canal stenosis. J Cell Biochem. https://doi.org/10.1002/jcb.28451
Hayashi K, Suzuki A, Terai H et al (2019) Fibroblast growth factor 9 is upregulated upon intervertebral mechanical stress-induced ligamentum flavum hypertrophy in a rabbit model. Spine 44:1172–1180. https://doi.org/10.1097/BRS.0000000000003089
Yabe Y, Hagiwara Y, Ando A et al (2015) Chondrogenic and fibrotic process in the ligamentum flavum of patients with lumbar spinal canal stenosis. Spine. https://doi.org/10.1097/BRS.0000000000000795
Pan B, Huo T, Cao M et al (2020) ADAM10 promotes the proliferation of ligamentum flavum cells by activating the PI3K/AKT pathway. Int J Mol Med 47:688–698. https://doi.org/10.3892/ijmm.2020.4809
Mori T, Sakai Y, Kayano M et al (2017) MicroRNA transcriptome analysis on hypertrophy of ligamentum flavum in patients with lumbar spinal stenosis. Spine Surg Relat Res 1:211–217. https://doi.org/10.22603/ssrr.1.2017-0023
Kong D, Zhao Q, Liu W, Wang F (2019) Identification of crucial miRNAs and lncRNAs for ossification of ligamentum flavum. Mol Med Rep. https://doi.org/10.3892/mmr.2019.10377
Wu W, Chen Y, Yang Z et al (2020) The role of gene expression changes in ceRNA network underlying ossification of ligamentum flavum development. DNA Cell Biol 39:1162–1171. https://doi.org/10.1089/dna.2020.5446
Xu Y, Zhang Z, Zheng Y, Feng S (2016) MicroRNA-221 regulates hypertrophy of ligamentum flavum in lumbar spinal stenosis by targeting TIMP-2. Spine 41:275–282. https://doi.org/10.1097/BRS.0000000000001226
Zhang B, Chen G, Yang X et al (2021) Dysregulation of microRNAs in hypertrophy and ossification of ligamentum flavum: new advances, challenges, and potential directions. Front Genet 12:641575. https://doi.org/10.3389/fgene.2021.641575
Han Y, Hong Y, Li L et al (2018) A transcriptome-level study identifies changing expression profiles for ossification of the ligamentum flavum of the spine. Mol Ther - Nucleic Acids 12:872–883. https://doi.org/10.1016/j.omtn.2018.07.018
Kolte VS, Khambatta S, Ambiye MV (2015) Thickness of the ligamentum flavum: correlation with age and its asymmetry-an magnetic resonance imaging study. Asian Spine J. https://doi.org/10.4184/asj.2015.9.2.245
Jung S-H, Young SS (2012) Power and sample size calculation for microarray studies. J Biopharm Stat 22:30–42. https://doi.org/10.1080/10543406.2010.500066
Huang DW, Sherman BT, Tan Q et al (2007) DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35:W169–W175. https://doi.org/10.1093/nar/gkm415
Szklarczyk D, Gable AL, Nastou KC et al (2021) The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612. https://doi.org/10.1093/nar/gkaa1074
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Isserlin R, Merico D, Voisin V, Bader GD (2014) Enrichment Map – a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. https://doi.org/10.12688/f1000research.4536.1
Bader GD, Hogue CWV (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2
Fabregat A, Sidiropoulos K, Viteri G et al (2017) Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 18:142. https://doi.org/10.1186/s12859-017-1559-2
Gillespie M, Jassal B, Stephan R et al (2022) The reactome pathway knowledgebase 2022. Nucleic Acids Res 50:D687–D692. https://doi.org/10.1093/nar/gkab1028
Viejo-Fuertes D, Liguoro D, Rivel J et al (1998) Morphologic and histologic study of the ligamentum flavum in the thoraco-lumbar region. Surg Radiol Anat. https://doi.org/10.1007/s00276-998-0171-6
Sairyo K, Biyani A, Goel V et al (2005) Pathomechanism of ligamentum flavum hypertrophy: a multidisciplinary investigation based on clinical, biomechanical, histologic, and biologic assessments. Spine. https://doi.org/10.1097/01.brs.0000188117.77657.ee
Sairyo K, Biyani A, Goel VK et al (2007) Lumbar ligamentum flavum hypertrophy is due to accumulation of inflammation-related scar tissue. Spine. https://doi.org/10.1097/01.brs.0000263407.25009.6e
Moon HJ, Kim JH, Park Y-K (2011) The angiogenic capacity from ligament flavum subsequent to inflammation: a critical component of the pathomechanism of hypertrophy. Spine J. https://doi.org/10.1016/j.spinee.2011.08.135
Yayama T, Uchida K, Kobayashi S et al (2007) Thoracic ossification of the human ligamentum flavum: histopathological and immunohistochemical findings around the ossified lesion. J Neurosurg Spine. https://doi.org/10.3171/SPI-07/08/184
Hirabayashi S (2017) Ossification of the ligamentum flavum. Spine Surg Relat Res 1:158–163. https://doi.org/10.22603/ssrr.1.2016-0031
Park JO, Lee BH, Kang YM et al (2013) Inflammatory cytokines induce fibrosis and ossification of human ligamentum flavum cells. J Spinal Disord Tech. https://doi.org/10.1097/BSD.0b013e3182698501
Zhang C, Chen Z, Meng X et al (2017) The involvement and possible mechanism of pro-inflammatory tumor necrosis factor alpha (TNF-α) in thoracic ossification of the ligamentum flavum. PLoS ONE. https://doi.org/10.1371/journal.pone.0178986
Chuang H-C, Tsai K-L, Tsai K-J et al (2020) Oxidative stress mediates age-related hypertrophy of ligamentum flavum by inducing inflammation, fibrosis, and apoptosis through activating Akt and MAPK pathways. Aging 12:24168–24183. https://doi.org/10.18632/aging.104105
Hur JW, Bae T, Ye S et al (2017) Myofibroblast in the ligamentum flavum hypertrophic activity. Eur Spine J 26:2021–2030. https://doi.org/10.1007/s00586-017-4981-2
Ornitz DM, Itoh N (2022) New developments in the biology of fibroblast growth factors. WIREs Mech Dis 14:e1549. https://doi.org/10.1002/wsbm.1549
Yun Y-R, Won JE, Jeon E et al (2010) Fibroblast growth factors: biology, function, and application for tissue regeneration. J Tissue Eng 1:218142. https://doi.org/10.4061/2010/218142
Lohmander LS, Hellot S, Dreher D et al (2014) Intraarticular sprifermin (recombinant human fibroblast growth factor 18) in knee osteoarthritis: a randomized, double-blind, placebo-controlled trial. Arthritis Rheumatol Hoboken NJ 66:1820–1831. https://doi.org/10.1002/art.38614
Fon Tacer K, Bookout AL, Ding X et al (2010) Research resource: comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol 24:2050–2064. https://doi.org/10.1210/me.2010-0142
Nakamura T, Okada T, Endo M et al (2014) Angiopoietin-like protein 2 induced by mechanical stress accelerates degeneration and hypertrophy of the ligamentum flavum in lumbar spinal canal stenosis. PLoS ONE. https://doi.org/10.1371/journal.pone.0085542
Habibi H, Suzuki A, Hayashi K et al (2022) Expression and function of fibroblast growth factor 1 in the hypertrophied ligamentum flavum of lumbar spinal stenosis. J Orthop Sci 27:299–307. https://doi.org/10.1016/j.jos.2021.01.004
Xie Y, Su N, Yang J et al (2020) FGF/FGFR signaling in health and disease. Signal Transduct Target Ther 5:181. https://doi.org/10.1038/s41392-020-00222-7
Flaherty KR, Wells AU, Cottin V et al (2019) Nintedanib in progressive fibrosing interstitial lung diseases. N Engl J Med 381:1718–1727. https://doi.org/10.1056/NEJMoa1908681
Burridge K, Guilluy C (2016) Focal adhesions, stress fibers and mechanical tension. Exp Cell Res 343:14–20. https://doi.org/10.1016/j.yexcr.2015.10.029
Liu L, Sui R, Li L et al (2022) Light activates Cdc42-mediated needle-shaped filopodia formation via the integration of small GTPases. Cell Mol Bioeng 15:599–609. https://doi.org/10.1007/s12195-022-00743-x
Ridley AJ, Allen WE, Peppelenbosch M, Jones GE (1999) Rho family proteins and cell migration. Biochem Soc Symp 65:111–123
Liang X, Xia R (2022) Kinesin family member 2A acts as a potential prognostic marker and treatment target via interaction with PI3K/AKT and RhoA/ROCK pathways in acute myeloid leukemia. Oncol Rep 47:18. https://doi.org/10.3892/or.2021.8229
McCormick B, Chu JY, Vermeren S (2019) Cross-talk between Rho GTPases and PI3K in the neutrophil. Small GTPases 10:187–195. https://doi.org/10.1080/21541248.2017.1304855
Acknowledgements
The authors thank Lorena Chávez, José Luis Santillán, Simón Guzmán, and Jorge Ramírez for the microarray analysis and Mayra Torres Quintana for her support in processing the tissues for histology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guevara-Villazón, F., Pacheco-Tena, C., Anchondo-López, A. et al. Transcriptomic alterations in hypertrophy of the ligamentum flavum: interactions of Rho GTPases, RTK, PIK3, and FGF. Eur Spine J 32, 1901–1910 (2023). https://doi.org/10.1007/s00586-023-07721-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-023-07721-5