Abstract
Background
Nodular gastritis (NG) is characterized by marked antral lymphoid follicle formation, and is a strong risk factor for diffuse-type gastric cancer in adults. However, it is unknown whether aberrant DNA methylation, which is induced by atrophic gastritis (AG) and is a risk for gastric cancer, is induced by NG. Here, we analyzed methylation induction by NG.
Methods
Gastric mucosal samples were obtained from non-cancerous antral tissues of 16 NG and 20 AG patients with gastric cancer and 5 NG and 6 AG patients without, all age- and gender-matched. Genome-wide methylation analysis and expression analysis were conducted by a BeadChip array and RNA-sequencing, respectively.
Results
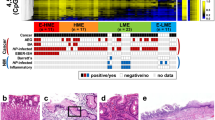
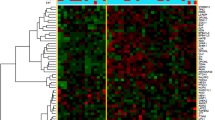
Clustering analysis of non-cancerous antral tissues of NG and AG patients with gastric cancer was conducted using methylation levels of 585 promoter CpG islands (CGIs) of methylation-resistant genes, and a large fraction of NG samples formed a cluster with strong methylation induction. Promoter CGIs of CDH1 and DAPK1 tumor-suppressor genes were more methylated in NG than in AG. Notably, methylation levels of these genes were also higher in the antrum of NG patients without cancer. Genes related to lymphoid follicle formation, such as CXCL13/CXCR5 and CXCL12/CXCR4, had higher expression in NG, and genes involved in DNA demethylation TET2 and IDH1, had only half the expression in NG.
Conclusions
Severe aberrant methylation, involving multiple tumor-suppressor genes, was induced in the gastric antrum and body of patients with NG, in accordance with their high gastric cancer risk.





Similar content being viewed by others
Abbreviations
- NG:
-
Nodular gastritis
- H. pylori :
-
Helicobacter pylori
- AG:
-
Atrophic gastritis
- CGIs:
-
CpG islands
- DNMTs:
-
DNA methyltransferases
- NO:
-
Nitric oxide
- FFPE:
-
Formalin-fixed paraffin-embedded
- FF:
-
Fresh-frozen
- NBI:
-
Narrow Band Imaging
- qPCR:
-
Quantitative PCR
- HSD:
-
Highest standard deviation
References
Ikuse T, Ohtsuka Y, Obayashi N, et al. Host response genes associated with nodular gastritis in Helicobacter pylori infection. Pediatr Int. 2018;60:446–54. https://doi.org/10.1111/ped.13527.
Nishikawa I, Kato J, Terasoma S, et al. Nodular gastritis in association with gastric cancer development before and after Helicobacter pylori eradication. JGH Open. 2018;2:80–6. https://doi.org/10.1002/jgh3.12049.
Okamoto K, Kodama M, Mizukami K, et al. Immunohistochemical differences in gastric mucosal damage between nodular and non-nodular gastritis caused by Helicobacter pylori infection. J Clin Biochem Nutr. 2021;69:216–21. https://doi.org/10.3164/jcbn.20-179.
Toyoshima O, Nishizawa T, Koike K. Endoscopic Kyoto classification of Helicobacter pylori infection and gastric cancer risk diagnosis. World J Gastroenterol. 2020;26:466–77. https://doi.org/10.3748/wjg.v26.i5.466.
Shiotani A, Kamada T, Kumamoto M, et al. Nodular gastritis in Japanese young adults: endoscopic and histological observations. J Gastroenterol. 2007;42:610–5. https://doi.org/10.1007/s00535-007-2073-5.
Asada K, Nakajima T, Shimazu T, et al. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut. 2015;64:388–96. https://doi.org/10.1136/gutjnl-2014-307094.
Maeda M, Nakajima T, Oda I, et al. High impact of methylation accumulation on metachronous gastric cancer: 5-year follow-up of a multicentre prospective cohort study. Gut. 2017;66:1721–3. https://doi.org/10.1136/gutjnl-2016-313387.
Maekita T, Nakazawa K, Mihara M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–95. https://doi.org/10.1158/1078-0432.Ccr-05-2096.
Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: the current state and clinical perspectives. Semin Cancer Biol. 2018;51:36–49. https://doi.org/10.1016/j.semcancer.2017.12.004.
Schneider BG, Mera R, Piazuelo MB, et al. DNA methylation predicts progression of human gastric lesions. Cancer Epidemiol Biomark Prev. 2015;24:1607–13. https://doi.org/10.1158/1055-9965.Epi-15-0388.
Yamashita S, Kishino T, Takahashi T, et al. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc Natl Acad Sci U S A. 2018;115:1328–33. https://doi.org/10.1073/pnas.1717340115.
Lee YC, Chiang TH, Chou CK, et al. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150:1113-24.e5. https://doi.org/10.1053/j.gastro.2016.01.028.
Usui G, Matsusaka K, Mano Y, et al. DNA methylation and genetic aberrations in gastric cancer. Digestion. 2021;102:25–32. https://doi.org/10.1159/000511243.
Kamada T, Tanaka A, Yamanaka Y, et al. Nodular gastritis with Helicobacter pylori infection is strongly associated with diffuse-type gastric cancer in young patients. Dig Endosc. 2007;19:180–4. https://doi.org/10.1111/j.1443-1661.2007.00750.x.
Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–9. https://doi.org/10.1038/ng892.
Takeshima H, Niwa T, Yamashita S, et al. TET repression and increased DNMT activity synergistically induce aberrant DNA methylation. J Clin Invest. 2020;130:5370–9. https://doi.org/10.1172/jci124070.
Takeuchi C, Sato J, Yamashita S, et al. Autoimmune gastritis induces aberrant DNA methylation reflecting its carcinogenic potential. J Gastroenterol. 2022. https://doi.org/10.1007/s00535-021-01848-2.
Kubes P, Jenne C. Immune responses in the liver. Annu Rev Immunol. 2018;36:247–77. https://doi.org/10.1146/annurev-immunol-051116-052415.
Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2011;14:101–12. https://doi.org/10.1007/s10120-011-0041-5.
Kaneko S, Yoshimura T. Time trend analysis of gastric cancer incidence in Japan by histological types, 1975–1989. Br J Cancer. 2001;84:400–5. https://doi.org/10.1054/bjoc.2000.1602.
Ueda S, Yamashita S, Watanabe SI, et al. Influence of degree of DNA degradation in formalin-fixed and paraffin-embedded tissue samples on accuracy of genome-wide DNA methylation analysis. Epigenomics. 2021;13:565–76. https://doi.org/10.2217/epi-2020-0431.
Iida N, Okuda Y, Ogasawara O, et al. MACON: a web tool for computing DNA methylation data obtained by the illumina infinium human DNA methylation BeadArray. Epigenomics. 2018;10:249–58. https://doi.org/10.2217/epi-2017-0093.
Yamashita S, Nanjo S, Rehnberg E, et al. Distinct DNA methylation targets by aging and chronic inflammation: a pilot study using gastric mucosa infected with Helicobacter pylori. Clin Epigenetics. 2019;11:191. https://doi.org/10.1186/s13148-019-0789-8.
Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7: e41361. https://doi.org/10.1371/journal.pone.0041361.
Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. https://doi.org/10.1093/bioinformatics/bts635.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. https://doi.org/10.1186/1471-2105-12-323.
Takeshima H, Yamashita S, Shimazu T, et al. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 2009;19:1974–82. https://doi.org/10.1101/gr.093310.109.
Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–8. https://doi.org/10.1038/bmt.2012.244.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. https://doi.org/10.1038/nprot.2008.211.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. https://doi.org/10.1093/nar/gkn923.
Song JZ, Stirzaker C, Harrison J, et al. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–61. https://doi.org/10.1038/sj.onc.1205153.
De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5’ region of gene MAGE-A1 in tumor cells. Mol Cell Biol. 2004;24:4781–90. https://doi.org/10.1128/mcb.24.11.4781-4790.2004.
Koide T, Koyanagi-Aoi M, Uehara K, et al. CDX2-induced intestinal metaplasia in human gastric organoids derived from induced pluripotent stem cells. iScience. 2022;25: 104314. https://doi.org/10.1016/j.isci.2022.104314.
Wang B, Wang M, Ao D, et al. CXCL13-CXCR5 axis: regulation in inflammatory diseases and cancer. Biochim Biophys Acta Rev Cancer. 2022;1877: 188799. https://doi.org/10.1016/j.bbcan.2022.188799.
Ferretti E, Ponzoni M, Doglioni C, et al. IL-17 superfamily cytokines modulate normal germinal center B cell migration. J Leukoc Biol. 2016;100:913–8. https://doi.org/10.1189/jlb.1VMR0216-096RR.
Hong W, Yang B, He Q, et al. New insights of CCR7 signaling in dendritic cell migration and inflammatory diseases. Front Pharmacol. 2022;13: 841687. https://doi.org/10.3389/fphar.2022.841687.
Yamamoto E, Suzuki H, Takamaru H, et al. Role of DNA methylation in the development of diffuse-type gastric cancer. Digestion. 2011;83:241–9. https://doi.org/10.1159/000320453.
Yuan W, Chen J, Shu Y, et al. Correlation of DAPK1 methylation and the risk of gastrointestinal cancer: a systematic review and meta-analysis. PLoS ONE. 2017;12: e0184959. https://doi.org/10.1371/journal.pone.0184959.
Grady WM, Willis J, Guilford PJ, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26:16–7. https://doi.org/10.1038/79120.
Guilford P, Hopkins J, Harraway J, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–5. https://doi.org/10.1038/32918.
Shin SH, Park SY, Ko JS, et al. Aberrant CpG island hypermethylation in pediatric gastric mucosa in association with Helicobacter pylori infection. Arch Pathol Lab Med. 2011;135:759–65. https://doi.org/10.5858/2010-0140-oa.1.
Choi E, Roland JT, Barlow BJ, et al. Cell lineage distribution atlas of the human stomach reveals heterogeneous gland populations in the gastric antrum. Gut. 2014;63:1711–20. https://doi.org/10.1136/gutjnl-2013-305964.
Kitamura S, Yasuda M, Muguruma N, et al. Prevalence and characteristics of nodular gastritis in Japanese elderly. J Gastroenterol Hepatol. 2013;28:1154–60. https://doi.org/10.1111/jgh.12180.
Acknowledgements
TU is a recipient of grants supported by AMED under Grant Nos. JP23ck0106804 and JP20gm1310006. HT is a recipient of a grant supported by JSPS KAKENHI under Grant No. JP21K07928. NY is a recipient of a Research Grant of the Princess Takamatsu Cancer Research Fund (ID: 19-25138) and JSPS KAKENHI under Grant No. JP21H03178.
Author information
Authors and Affiliations
Contributions
AS, HT, and TU conceived and designed the study. AS, HT, and YO carried out experiments. AS, HT, SY, and AH conducted data analysis. CI, JK, CT, MF, YF, NY, TA, HK, TK, CS, and KK collected clinical samples and WN supervised the pathological diagnosis and sample processing. AS, HT, TI, and TU wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sasaki, A., Takeshima, H., Yamashita, S. et al. Severe induction of aberrant DNA methylation by nodular gastritis in adults. J Gastroenterol (2024). https://doi.org/10.1007/s00535-024-02094-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00535-024-02094-y