Abstract
Background
Upregulated Kindlin-2 expression in hepatocellular carcinoma (HCC) correlates with metastasis and poor prognosis. In this study, we investigated the molecular mechanism of Kindlin-2 in HCC.
Methods
Kindlin-2 downstream pathways were explored through microRNA sequencing. The Kindlin-2–miR-1258–TCF4 axis was verified using bisulfite sequencing, a luciferase reporter assay, quantitative real-time PCR, and rescue assays. Binding of TCF4 to the Kindlin-2 promoter was confirmed by promoter activity analysis and chromatin immunoprecipitation.
Results
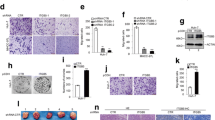
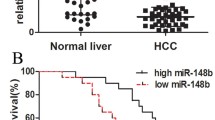
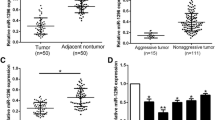
MiRNA sequencing identified miR-1258 as a downstream effector of Kindlin-2. MiR-1258 expression was increased following Kindlin-2 knockdown and decreased after Kindlin-2 overexpression. Next, we identified transcription factor 7 like 2 (TCF7L2 or TCF4) as a target of miR-1258 and found that Kindlin-2 upregulated TCF4 expression by epigenetically suppressing miR-1258 in HCC. Furthermore, our results suggest that TCF4 binds to the Kindlin-2 promotor to enhance its transcription. Therefore, Kindlin-2–miR-1258–TCF4 interaction creates a positive feedback loop. Functional assays and animal experiments demonstrated critical roles of miR-1258 and TCF4 in HCC cell migration in vitro and HCC metastasis in vivo. In HCC tissues, Kindlin-2 expression correlated negatively with miR-1258 expression and positively with TCF4 expression. Meanwhile, miR-1258 expression correlated negatively with TCF4 expression.
Conclusions
This study illustrates a novel integrin-independent signaling pathway, Kindlin-2–miR-1258–TCF4, that regulates HCC invasion and metastasis and identifies Kindlin-2 as a promising therapeutic target in HCC.







Similar content being viewed by others
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- EMT:
-
Epithelial–mesenchymal transition
- miR:
-
MicroRNA
- SDS-PAGE:
-
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- shRNA:
-
Short hairpin RNAs
- CPM:
-
Counts per million reads
- qRT-PCR:
-
Quantitative real-time PCR
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- Cas 9:
-
CRISPR-associated protein 9
- ChIP:
-
Chromatin immunoprecipitation
- DNMT:
-
DNA methyltransferase
- 5-aza-CdR:
-
5-Aza-2’-deoxycytidine
- TCF4:
-
Transcription factor 7 like 2
- FERM:
-
4.1-Ezrin-ridixin-moesin
- 3′-UTR:
-
3′-Untranslated region
- CDS:
-
Coding sequence
References
Li H, Deng Y, Sun K, et al. Structural basis of kindlin-mediated integrin recognition and activation. Proc Natl Acad Sci USA. 2017;114:9349–54.
Wang W, Kansakar U, Markovic V, et al. Role of Kindlin-2 in cancer progression and metastasis. Ann Transl Med. 2020;8:901.
Lin J, Lin W, Ye Y, et al. Kindlin-2 promotes hepatocellular carcinoma invasion and metastasis by increasing Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2017;36:134.
Zhang HF, Alshareef A, Wu C, et al. Loss of miR-200b promotes invasion via activating the Kindlin-2/integrin β1/AKT pathway in esophageal squamous cell carcinoma: an E-cadherin-independent mechanism. Oncotarget. 2015;6:28949–60.
Shen Z, Ye Y, Kauttu T, et al. Novel focal adhesion protein kindlin-2 promotes the invasion of gastric cancer cells through phosphorylation of integrin β1 and β3. J Surg Oncol. 2013;108:106–12.
Yu Y, Wu J, Guan L, et al. Kindlin 2 promotes breast cancer invasion via epigenetic silencing of the microRNA200 gene family. Int J Cancer. 2013;133:1368–79.
Ren W, Gao L, Qiang C, et al. Kindlin-2-mediated upregulation of ZEB2 facilitates migration and invasion of oral squamous cell carcinoma in a miR-200b-dependent manner. Am J Transl Res. 2018;10:2529–41.
Zhang HF, Zhang K, Liao LD, et al. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting Kindlin-2. Carcinogenesis. 2014;35:292–301.
Sossey-Alaoui K, Pluskota E, Szpak D, et al. The Kindlin-2 regulation of epithelial-to-mesenchymal transition in breast cancer metastasis is mediated through miR-200b. Sci Rep. 2018;8:7360.
Sossey-Alaoui K, Pluskota E, Bialkowska K, et al. Kindlin-2 regulates the growth of breast cancer tumors by activating CSF-1-mediated macrophage infiltration. Cancer Res. 2017;77:5129–41.
Wang Z, Yang Y, Cui Y, et al. Tumor-associated macrophages regulate gastric cancer cell invasion and metastasis through TGFβ2/NF-κB/Kindlin-2 axis. Chin J Cancer Res. 2020;32:72–88.
Guo B, Gao J, Zhan J, et al. Kindlin-2 interacts with and stabilizes EGFR and is required for EGF-induced breast cancer cell migration. Cancer Lett. 2015;361:271–81.
Gao J, Khan AA, Shimokawa T, et al. A feedback regulation between Kindlin-2 and GLI1 in prostate cancer cells. FEBS Lett. 2013;587:631–8.
Peng W, Wen HC, Xi Z, et al. Kindlin-2 interacts with and stabilizes DNMT1 to promote breast cancer development. Int J Biochem Cell Biol. 2018;105:41–51.
Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6.
Ge YS, Liu D, Jia WD, et al. Kindlin-2: a novel prognostic biomarker for patients with hepatocellular carcinoma. Pathol Res Pract. 2015;211:198–202.
Sun Z, Costell M, Fässler R. Integrin activation by talin, kindlin and mechanical forces. Nat Cell Biol. 2019;21:25–31.
Zhan J, Zhang H. Kindlins: roles in development and cancer progression. Int J Biochem Cell Biol. 2018;98:93–103.
Hu M, Wang M, Lu H, et al. Loss of miR-1258 contributes to carcinogenesis and progression of liver cancer through targeting CDC28 protein kinase regulatory subunit 1B. Oncotarget. 2016;7:43419–31.
Huang WJ, Tian XP, Bi SX, et al. The β-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene. 2020;39:4538–50.
Zou H, Xu X, Luo L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019;18:2398–413.
Fang Q, Liu H, Zhou A, et al. Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manag Res. 2020;12:6849–60.
Li W, Yang X, Shi C, et al. Hsa_circ_002178 promotes the growth and migration of breast cancer cells and maintains cancer stem-like cell properties through regulating miR-1258/KDM7A axis. Cell Transplant. 2020;29:963689720960174.
Zhang W, Wu G, Sun P, et al. circ_SMAD2 regulate colorectal cancer cells proliferation through targeting miR-1258/RPN2 signaling pathway. J Cancer. 2021;12:1678–86.
Wang LQ, Kumar S, Calin GA, et al. Frequent methylation of the tumour suppressor miR-1258 targeting PDL1: implication in multiple myeloma-specific cytotoxicity and prognostification. Br J Haematol. 2020;190:249–61.
Loginov VI, Burdennyy AM, Filippova EA, et al. Hypermethylation of miR-107, miR-130b, miR-203a, miR-1258 genes associated with ovarian cancer development and metastasis. Mol Biol. 2018;52:801–9.
Zheng DL, Zhang L, Cheng N, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol. 2009;50:377–87.
Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20.
Caspi M, Wittenstein A, Kazelnik M, et al. Therapeutic targeting of the oncogenic Wnt signaling pathway for treating colorectal cancer and other colonic disorders. Adv Drug Deliv Rev. 2021;169:118–36.
Yu Y, Wu J, Wang Y, et al. Kindlin 2 forms a transcriptional complex with β-catenin and TCF4 to enhance Wnt signalling. EMBO Rep. 2012;13:750–8.
Wedel M, Fröb F, Elsesser O, et al. Transcription factor Tcf4 is the preferred heterodimerization partner for Olig2 in oligodendrocytes and required for differentiation. Nucleic Acids Res. 2020;48:4839–57.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant number: 81802882), Joint Funds for the innovation of Science and Technology, Fujian province (Grant number: 2018Y9104, 2018Y9108), and the Natural Science Foundation of Fujian Province (Grant number: 2018J01251).
Funding
National Natural Science Foundation of China, 81802882, Jie Lin, Joint Funds for the innovation of Science and Technology, Fujian province, 2018Y9104, Wansong Lin, 2018Y9108, Yunbin Ye, Natural Science Foundation of Fujian Province, 2018J01251, Jie Lin.
Author information
Authors and Affiliations
Contributions
LWS and LJ conceived and designed the study as well as performed the laboratory analysis. LY and WYJ performed the animal experiments and the ChIP analysis. LJY and CSP performed the western blotting analysis. CLF performed the qRT-PCR analysis in clinical samples. CH and LL collected the samples. LJ, LWS, CXY, and YYB contributed reagents, materials, and analysis tools. LWS and LJ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.



Rights and permissions
About this article
Cite this article
Lin, W., Lin, J., Li, J. et al. Kindlin-2–miR-1258–TCF4 feedback loop promotes hepatocellular carcinoma invasion and metastasis. J Gastroenterol 57, 372–386 (2022). https://doi.org/10.1007/s00535-022-01866-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-022-01866-8