Abstract
Background
Intraepithelial lymphocytes (IELs) in the intestine play important roles in the regulation of local immune responses. Although their functions have been studied in a variety of animal experiments, in vitro studies on spatiotemporal behaviors of IELs and their interaction with intestinal epithelial cells (IECs) have been hampered due to the lack of a suitable culture system. In this study, we aimed at developing a novel co-culture system of IELs with IECs to investigate dynamic interaction between these two populations of cells in vitro.
Methods
We optimized experimental conditions under which murine IELs can be efficiently maintained with IECs cultured as three-dimensional organoids. We then tested the effect of IL-2, IL-7, and IL-15 on the maintenance of IELs in this co-culture system. By time-lapse imaging, we also examined the dynamic behaviors of IELs.
Results
IELs can be expanded with epithelial organoids in the presence of IL-2, IL-7, and IL-15. IELs were efficiently maintained within and outside of organoids showing a ~four-fold increase in both αβT and γδT IELs for a period of 2 weeks. Four-dimensional fluorescent imaging revealed an active, multi-directional movement of IELs along the basolateral surface of IECs, and also their inward or outward migration relative to organoid structures. Cell tracking analysis showed that αβT and γδT IELs shared indistinguishable features with regard to their dynamics.
Conclusions
This novel co-culture method could serve as a unique tool to investigate the motility dynamics of IELs and their temporal and spatial interaction with IECs in vitro.
Similar content being viewed by others
Introduction
In the intestine, the epithelial layer covering its inner-most surface functions as not only the site of substance passage but also the active site of immune response [1, 2]. The epithelium consists of different types of intestinal epithelial cells (IECs) that arise from adult stem cells residing at the bottom of the crypt-villus epithelial architecture. The intestinal epithelial tissue is also home to an abundant population of cells of a different lineage, intestinal intraepithelial lymphocytes (IELs). For example, IELs are scattered at a density of one IEL per 5–10 IECs in the small intestine [3], making intimate contact with IECs.
IELs are heterogeneous populations that contain T cell receptor (TCR) αβ T cells (αβT IELs) and TCR γδ T cells (γδT IELs) [4]. Among αβT IELs, CD4 + CD8αβ-CD8αα+, CD4 + CD8αβ-CD8αα−, and CD4-CD8αβ+ subsets are classified as conventional IELs, as they develop through conventional thymic selection and migrate into the intestine [5]. Other αβT IELs of the CD4-CD8αβ− double negative (DN) phenotype (CD4-CD8αβ-CD8αα+ and CD4-CD8αβ-CD8αα- cells) and γδT IELs are classified as unconventional subsets, as they develop and become activated differently from conventional IELs [5, 6]. Although both αβT and γδT IELs are known to play important roles in local immune response [7–10], functions of IELs in each subset are not fully characterized.
As IELs are highly susceptible to apoptosis when isolated [11–13], in vitro study of IEL functions has long been hampered. This has been attributed to the lack of some IEC-derived factors important for survival and proliferation of IELs in in vitro culture systems. Indeed, the supply of factors derived from IECs [13, 14] or by direct contact with laboratory-adapted epithelial cell lines [15–17] was shown to be beneficial for in vitro maintenance of IELs. This supports the idea that IECs that preserve physiological properties would provide a more suitable microenvironment for sustained culture of IELs.
Recent advances have enabled long-term culture of IECs in vitro [18–20]. For example, with such a technology, IECs of the murine small intestine are now allowed to grow almost in perpetuity as three-dimensional epithelial organoids [18]. IECs expanded by this method are capable of reconstituting normal epithelia when transplanted back into syngeneic mice, indicating that the culture process is not deleterious to maintenance of their original features [21]. Thus, it would be of interest to investigate whether the organoid culture technology could become a novel platform to achieve efficient co-culture systems for IELs. In addition, such co-culture systems could offer the advantages of a physiologically relevant model system to study functional and/or temporal and spatial interaction between these two cell populations.
Methods
Mice
EGFP transgenic mice [22], R26-H2B-EGFP mice, and R26-H2B-mCherry mice [23] of C57BL6 background were bred and maintained in the animal facility of Tokyo Medical and Dental University (TMDU). All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of TMDU.
Preparation of intestinal organoids
Small intestinal crypts were isolated from 10 to 15-week-old wild-type mice as previously described [18]. The crypts were embedded in Matrigel (BD Biosciences) and Advanced DMEM/F12 containing 500 ng/mL mRspo1 (R&D Systems), 20 ng/mL mEGF (Peprotech), and 100 ng/mL mNoggin (R&D Systems) were added to each well. The medium was changed every 2 days until the following use of cultured organoids.
Isolation of IELs
Intestinal IELs were isolated according to the method described previously [24]. The small intestine of 10–25-week-old mice was incubated in a tube containing 5 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM 1,4-dithio-d-threitol (DTT) in Ca- and Mg-free Hank’s balanced salt solution. After passage of the cell suspension through a nylon mesh filter and a column of glass wool, recovered IELs were collected by Percoll gradient centrifugation. After washing, cells were counted and used immediately for the following assays. When needed, αβT and γδT IELs were sorted using a FACS ARIA II (Becton–Dickinson) by negative selection. Antibodies used were as follows: anti-mouse TCR-β-PE (BioLegend) and anti-mouse TCR-δ-PECy7 (BioLegend). Purity of sorted IEL subsets was analyzed after sorting and was always greater than 99 %.
Co-culture of IELs and IECs
Intestinal organoids were cultured for 2 days prior to the co-culture with IELs. On the day of IEL isolation, cultured organoids were released from Matrigel by incubating whole Matrigel in cell recovery solution (Corning) on ice for 30 min. After washing and counting, organoids and independently isolated IELs were placed together onto a 24-well plate, resulting in 100 organoids and 1.0 × 105 IELs were intermixed with 400 μL of DMEM. After incubation at 37 °C for 30 min, the mixture was gently collected and centrifuged for 1 min at 200g. The pellet was suspended in 30 μL of Matrigel and placed in 24-well plates. After Matrigel polymerization, 500 μL of the same medium as that used for the organoid culture was added to each well. Where indicated, 100 U/mL recombinant human IL-2 (Roche), 10 ng/mL mouse IL-7 (Peprotech) and 10 ng/mL mouse IL-15 (Peprotech) were supplemented to the co-culture. The medium was refreshed every 2 days.
After 7 days of co-culture, IELs could be propagated as follows. Entire cells were released from Matrigel by Cell Recovery Solution as described previously [18]. After centrifugation at 500g for 5 min, supernatants were collected and stored as fraction 1. To facilitate recovery of IELs still incorporated in organoids, the pellet was incubated in 5 mM EDTA/PBS for 10 min on ice. The tube was left for 1 min so that the organoids sedimented under gravity, and the supernatant collected and stored as fraction 2. To recover more IELs, sedimented materials were mechanically disrupted by vigorous pipetting and then passed through a 40 μm nylon mesh filter (fraction 3). Fractions 1–3 were combined and EGFP + IELs were counted by a hemocytometer. In parallel, intestinal crypts were freshly isolated as a counterpart of co-culture. Recovered IELs and isolated organoids were mixed as described previously, and co-cultured.
Immunohistochemistry and fixed cell analysis
For whole-mount analysis, cells were fixed with surrounding Matrigel and permeabilized. Primary antibodies were incubated overnight and secondary antibody reactions were performed for 1.5 h. Nuclei were stained with 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI). Samples were then overlaid with Vectashield Mounting Medium (Vector Laboratories) and microscopically analyzed. Antibodies used were as follows: CD3ε (Becton–Dickinson) and Cdh1 (Santa Cruz Biotechnology). Confocal imaging was performed on a Fluoview FV10i system (Olympus). Images were taken using an Olympus 60× objective (1.35 N.A.) at Z-steps of 0.85 μm. The Z-stacks of images were processed with Adobe Photoshop software when necessary.
Flow cytometry
IELs were suspended in 0.2 % FCS/PBS. Cells were analyzed on a FACS Canto II (BD Biosciences). Antibodies used for surface marker analysis were as follows: anti-mouse TCR-β-PE (BioLegend) and anti-mouse TCR-δ-PECy7 (BioLegend). For intracellular staining, IELs were fixed, permeabilized with PermI reagents (Becton–Dickinson) and incubated with anti-Ki-67-660eFluor (eBioscience) antibodies.
Time-lapse fluorescent imaging
Time-lapse imaging of the co-culture was performed on a DeltaVision system (Applied Precision) in which a fluorescent microscope IX-71 (Olympus) with an InsightSSI illumination source is incorporated. Before initiation of imaging, cell nuclei were stained with Hoechst 33342 (Nacalai Tesque). Fluorescent images of nuclei, EGFP, and mCherry were acquired through a UplansApo 20x objective (0.75 N.A.) or a UApo340 40x objective (1.34 N.A.) onto a CoolSnap ES2 digital camera (Roper Scientific). Single-plane imaging was performed for 2 h at 20 s intervals. Multi-plane imaging was conducted for 10 min at 20 or 30 s intervals, acquiring Z-stacks at 5 μm steps at a time. SoftWorx software (Applied Precision) was used to obtain maximum intensity projection data of these Z-stacks.
IEL motility analysis
Datasets were imported into Imaris 7.5 (Bitplane) software. Nuclei of IELs were then individually determined by using the spot-tracking feature of Imaris, with a size estimate of 10 µm. The migration of nuclei was analyzed by the “autoregressive motion” tracking algorithm of the software. Tracks were also verified visually to ensure that one track followed the same nucleus for the entire time period. The calculations of the mean speed, maximum speed, track length, and displacement of nuclei were performed using the Imaris software. Data are presented as mean ± S.E.M.
Results
Development of 3D co-culture of IELs and intestinal organoids
As a preliminary experiment to develop a method to maintain IELs with intestinal organoids, we assessed whether the conventional organoid culture allowed concomitantly isolated IELs to remain incorporated. Murine small intestinal crypts were isolated, embedded in Matrigel, and cultured as described previously [18]. The organoids were fixed together with surrounding Matrigel at various time points and immunostained three-dimensionally. This whole-mount analysis revealed that there were no detectable CD3+ cells in any organoids analyzed on day 3 or 7 (data not shown). This showed that the previously reported method of intestinal organoid culture does not allow IELs to be efficiently maintained.
We thus designed an experimental system wherein separately isolated IELs and IECs were cultured together. IELs were isolated from EGFP-tg mice of C57BL6 background. Meanwhile, wild-type IECs were cultured as organoids for 2 days and recovered from Matrigel. These two populations of cells were mixed and co-cultured in fresh Matrigel (Fig. 1a). Whole-mount analysis 2 days after starting co-culture (day 3) showed that EGFP + IELs were assembled into organoids (Fig. 1a). EGFP+ cells were positive for CD3 expression, indicating preservation of their T cell phenotype (Fig. 1a). Z-stack analysis showed that all organoids included at least one EGFP+ cell (100 %). The number of EGFP + IELs was 6.9 ± 0.8 per organoid on day 3 (n = 24). Interestingly, EGFP + IELs were present not only in organoids, but also external to these structures without any obvious physical contact (Fig. 1a). Previous studies showed that the in vitro life span of isolated IELs, irrespective of their TCRαβ and TCRγδ phenotypes, is limited to a period of less than 48 h in the absence of cytokine supplementation or TCR stimulation [25, 26]. Thus, the presence of these seemingly reclusive IELs on day 3 suggested that, for their prolonged survival, there might be some supportive functions exerted distantly by IECs existing in the same microenvironment. When analyzed on day 7, EGFP+ cells became sparse in both organoids and inter-organoid space (Fig. 1b). This indicated that co-culture of separately isolated IELs and IECs allows short-term maintenance of IELs, although further optimization is still possible.
Cultured intestinal organoids permit in vitro maintenance of IELs. a IELs and IECs were separately isolated from EGFP-tg mice and wild-type mice, respectively. The cells were co-cultured and whole-mount analysis was performed on day 3. Three representative images along the Z-stacks of an organoid are shown with immunostaining of CD3 (red) and endogenous EGFP (green) with nuclear staining. Arrowheads show IELs incorporated into the organoid, and arrows show those existing externally to organoids. b Time course changes of co-culture (days 1, 3 and 7) are shown. Merged images of EGFP signal (IELs) and phase-contrast (PC) view are shown. Scale bars show 50 μm
IL-2, IL-7, and IL-15 are known to prolong the in vitro life span of αβT and γδT IELs through multiple mechanisms [13, 14, 25, 27]. To test whether these cytokines also extend culture longevity of IELs in our system, co-culture was supplemented with IL-2, IL-7, IL-15, or a combination of all three cytokines. As the size of organoids became too large to perform whole-mount imaging at day 7 or thereafter, we collected EGFP+ cells as single cells and counted them using flow cytometry. The number of EGFP + cells was below the detectable level in the absence of cytokines (Fig. 2a). By contrast, IL-7 and IL-15 had positive effects on survival and/or growth of IELs. IL-2 showed more potent effects than IL-7 or IL-15, resulting in an increase in cell number during co-culture (Fig. 2a). The IEL count significantly increased (two- to three-fold) when the culture was supplemented with the mixture of IL-2/-7/-15, suggesting their additive effects (Fig. 2a, b). Of note, when EGFP + IELs were re-isolated on day 7 and co-cultured with freshly prepared epithelial organoids in the presence of IL-2/-7/-15, they could be further expanded for at least one more week, resulting in an approximately four-fold increase over the entire culture period (Fig. 2b). Expansion of IELs by cytokine addition was also confirmed by microscopy. We found the increase of two populations of EGFP + IELs in culture on day 7—those associated with intestinal organoids and those maintained outside the organoids (Fig. 2c).
IL-2/-7/-15 enhances expansion of αβT and γδT IELs. a IELs from EGFP-tg mice (10 × 104 per well) and wild-type IECs were co-cultured in the presence of either 100 U/mL of IL-2, 10 ng/mL of IL-7, 10 ng/mL of IL-15, or all three cytokines (IL-2/-7/-15). IELs were recovered and counted on day 7. b EGFP + IELs (10 × 104 per well) were cultured with IECs as in a in the presence of IL-2/-7/-15. Numbers of IELs were counted on day 1 (before co-culture), day 7 and 14. In a and b, the values were expressed as the number of cells per well ± SEM in three independent experiments. c Time course changes of co-culture (day 1, 3 and 7) supplemented with IL-2/-7/-15. EGFP signals (IELs) and their merged images with phase-contrast (PC) view are shown. High-power views of the area within dotted squares are shown at the bottom. Scale bars show 100 μm. d, e IELs on day 1 (before culture) and day 7 were analyzed by flow cytometry for their surface expression of TCRαβ and TCRγδ (d), or intracellular expression of Ki-67 (e). Representative data from three independent experiments for IELs from different donors are shown
We next assessed whether αβT and γδT IELs were both maintained in culture. Flow cytometry analysis revealed that proportions of these subtypes in the whole population were comparable before and after culture (day 7) (Fig. 2d). αβT and γδT IELs in culture showed higher Ki-67 expression compared to those analyzed before culture, indicating the presence of proliferating cells in both subpopulations (Fig. 2e). Similar data were obtained in IELs propagated for 2 weeks (Suppl. Fig. 1).
Motility analysis of IELs in the co-culture system
We then analyzed the temporal and spatial interaction between IELs and organoids by using this novel co-culture system. After 2 days of co-culture, EGFP + IELs and wild-type organoids were subjected to single-plane time-lapse imaging. Interestingly, most IELs were found to be highly motile throughout the observation time periods (Fig. 3a and Suppl. movie 1). We were able to document that some EGFP + IELs migrated toward the organoid, came in direct contact with it, stayed temporarily in it, and then egressed from it (Fig. 3a, top). IELs associating with the organoid were also motile. They migrated along the basal side of the epithelial monolayer in a random direction (Fig. 3a, bottom). To investigate this more in detail, we performed 4-D imaging analysis using epithelial organoids obtained from histone H2B-mCherry mice [23], which allowed for visualization of nuclei of IECs (Fig. 3b and Suppl. Movie 2). We were able to observe that IELs continually and extensively change their cellular shape over time, occasionally interposing their cytoplasmic projections between two nuclei of adjacent epithelial cells (Fig. 3b and Suppl. Movie 2).
Dynamic migration of IELs co-cultured with intestinal organoids. a Single-plane time-lapse imaging of co-cultured EGFP + IELs and wild-type IECs performed for 2 h at 20 s intervals. The view at the start of imaging is shown on the left (t = 0:00:00) with nuclei stained using Hoechst 33342. Representative cells migrating inward and outward from the organoid (top), and those moving along its basal side (bottom) are shown. Whole images can be viewed as Suppl. Movie 1. b 4D imaging to track IEL movement. IELs from EGFP-tg mice and IECs from histone H2B-mCherry mice were co-cultured. On day 3, multi-plane time-lapse imaging was performed for 20 min at 20 s intervals. Serial images of a representative cell (arrowhead) are shown on the right. Whole images can be viewed as Suppl. Movie 2
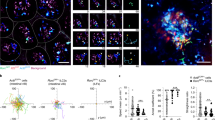
We next attempted to quantitatively analyze the migration activities of IELs. In order to track individual cells precisely, we used IELs isolated from histone H2B-EGFP mice [23] (Fig. 4a) for this experiment. EGFP + IELs were co-cultured with wild-type organoids, and imaging was performed acquiring around 20 Z-stack images at a time. The maximum intensity projection of Z-stacked images showed that the nuclei of individual IELs were clearly detectable with this approach (Fig. 4b). Again, we could observe IELs migrating along the epithelial layer of organoids, and also those migrating into and out of the organoid dynamically. By reconstructing three-dimensional data, we measured migration parameters of IELs. The mean and maximum speed values of their migration were 5.07 ± 0.27 and 9.67 ± 0.60 μm/min, respectively (Fig. 4c; Table 1 and Suppl. Movie 3). To test whether αβT and γδT IELs differ in their migratory properties, separately isolated αβT and γδT IELs were co-cultured with organoids and their dynamic behaviors were similarly assessed. There was no significant difference between the two populations in any of the parameters obtained (Fig. 4d; Table 1). Comparable data were also obtained by analysis of IELs cultured for 2 weeks (Table 1).
Quantitative analysis of IEL motility in co-culture system. a 4D imaging was used to quantitatively assess IEL migration. IELs from H2B-EGFP mice and IECs from wild-type mice were co-cultured. On day 3, multi-plane time-lapse imaging was performed for 10 min at 30 s intervals. Nuclei that stayed inside the inner area within the data acquisition space for more than 80 % of the observation period were analyzed. b A still image of the representative movie is shown as the maximum intensity projection of Z-stacks (left). Cells migrating along the organoid (top), and those showing inward and outward migration (bottom) are shown. c Three-dimensional cell tracking analysis. Nuclei of IELs (yellow spheres) and their trajectories (white lines) are shown (left). A magnified image of the boxed square is also shown (right). Whole data are available as Suppl. Movie 3. d αβT and γδT IELs were separately isolated from H2B-EGFP reporter mice and co-cultured with the organoids of wild-type mice. On day 3 of co-culture, time-lapse imaging and cell tracking analyses were performed as in c. Still images cropped from representative movies are shown. Indicated time denotes time after start of each imaging. Scale bars show 10 μm
Discussion
We have developed a novel method to culture murine IELs in vitro in which both αβT and γδT IELs proliferate efficiently in the presence of epithelial organoids. Based on the previous observations that IEC-derived factors [13, 14] or direct contact with epithelial cell lines [5, 16, 17] did not allow for efficient expansion of IELs, we assume that non-transformed IEC-derived factors and their three-dimensional presentation to IELs may be important for the maintenance of IELs in our system. Although such niche factors remain to be identified, the culture method presented in this study could serve as a useful tool for the behavioral characterization of IELs.
By combining time-lapse imaging with the culture system, we also showed that IELs move around three-dimensionally with high motility, changing their contact status with epithelial organoids. A previous study assessed motility parameters of γδT IELs using in vivo confocal microscopy and determined the mean and maximum velocities as 3.8 ± 0.1 and 7.7 μm/min, respectively [28]. As quantitative analysis for the motility of both αβT and γδT IELs showed comparable values in our analysis, it was proposed that IELs in this in vitro system are as motile as IELs residing in in vivo settings.
In summary, we have demonstrated a novel co-culture method to maintain and expand murine IELs. Using this in vitro culture system, we also demonstrated the highly dynamic nature of IELs. The method will serve as a unique tool to investigate functions and dynamics of IELs that interact with IECs in vivo.
References
Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809.
Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–53.
Beagley KW, Fujihashi K, Lagoo AS, et al. Differences in intraepithelial lymphocyte T cell subsets isolated from murine small versus large intestine. J Immunol. 1995;154:5611–9.
Goodman T, Lefrançois L. Expression of the gamma-delta T-cell receptor on intestinal CD8+ intraepithelial lymphocytes. Nature. 1988;333:855–8.
Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–56.
Hayday A, Theodoridis E, Ramsburg E, et al. Intraepithelial lymphocytes: exploring the third way in immunology. Nat Immunol. 2001;2:997–1003.
Boismenu R, Havran WL. Modulation of epithelial cell growth by intraepithelial gamma delta T cells. Science. 1994;266:1253–5.
Chen Y, Chou K, Fuchs E, et al. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci USA. 2002;99:14338–43.
Müller S, Bühler-Jungo M, Mueller C. Intestinal intraepithelial lymphocytes exert potent protective cytotoxic activity during an acute virus infection. J Immunol. 2000;164:1986–94.
Masopust D, Vezys V, Marzo AL, et al. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–7.
Viney JL, MacDonald TT. Selective death of T cell receptor gamma/delta+ intraepithelial lymphocytes by apoptosis. Eur J Immunol. 1990;20:2809–12.
Yada S, Kishihara K, Kong YY, et al. Differential requirements of CD45 protein tyrosine phosphatase for cytolytic activities and intrathymic and extrathymic development of intestinal intraepithelial lymphocytes. J Immunol. 1998;161:2208–16.
Chu CL, Chen SS, Wu TS, et al. Differential effects of IL-2 and IL-15 on the death and survival of activated TCR gamma delta+ intestinal intraepithelial lymphocytes. J Immunol. 1999;162:1896–903.
Inagaki-Ohara K, Nishimura H, Mitani A, et al. Interleukin-15 preferentially promotes the growth of intestinal intraepithelial lymphocytes bearing gamma delta T cell receptor in mice. Eur J Immunol. 1997;27:2885–91.
Chennupati V, Worbs T, Liu X, et al. Intra- and intercompartmental movement of gammadelta T cells: intestinal intraepithelial and peripheral gammadelta T cells represent exclusive nonoverlapping populations with distinct migration characteristics. J Immunol. 2010;185:5160–8.
Zufferey C, Erhart D, Saurer L, et al. Production of interferon-gamma by activated T-cell receptor-alphabeta CD8alphabeta intestinal intraepithelial lymphocytes is required and sufficient for disruption of the intestinal barrier integrity. Immunology. 2009;128:351–9.
Bas A, Swamy M, Abeler-Dörner L, et al. Butyrophilin-like 1 encodes an enterocyte protein that selectively regulates functional interactions with T lymphocytes. Proc Natl Acad Sci USA. 2011;108:4376–81.
Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5.
Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72.
Yui S, Nakamura T, Sato T, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. 2012;18:618–23.
Fukuda M, Mizutani T, Mochizuki W, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. 2014;28:1752–7.
Okabe M, Ikawa M, Kominami K, et al. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9.
Abe T, Kiyonari H, Shioi G, et al. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis. 2011;49:579–90.
Kohyama M, Nanno M, Kawaguchi-Miyashita M, et al. Cytolytic and IFN-gamma-producing activities of gamma delta T cells in the mouse intestinal epithelium are T cell receptor-beta-chain dependent. Proc Natl Acad Sci USA. 1999;96:7451–5.
Lai YG, Gelfanov V, Gelfanova V, et al. IL-15 promotes survival but not effector function differentiation of CD8+ TCRalphabeta+ intestinal intraepithelial lymphocytes. J Immunol. 1999;163:5843–50.
Brunner T, Arnold D, Wasem C, et al. Regulation of cell death and survival in intestinal intraepithelial lymphocytes. Cell Death Differ. 2001;8:706–14.
Yada S, Nukina H, Kishihara K, et al. IL-7 prevents both caspase-dependent and-independent pathways that lead to the spontaneous apoptosis of i-IEL. Cell Immunol. 2001;208:88–95.
Edelblum KL, Shen L, Weber CR, et al. Dynamic migration of γδ intraepithelial lymphocytes requires occludin. Proc Natl Acad Sci USA. 2012;109:7097–102.
Acknowledgments
We thank Lesa Thompson for manuscript editing. This study was supported by MEXT Kakenhi (Grant Number 26112705), JSPS Kakenhi (24390186, 24590936, 26221307), and Health and Labour Sciences Research Grants for research on intractable diseases from Ministry of Health, Labor and Welfare of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nozaki, K., Mochizuki, W., Matsumoto, Y. et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol 51, 206–213 (2016). https://doi.org/10.1007/s00535-016-1170-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-016-1170-8