Abstract
Background
Resistance-associated variants (RAVs) reduce the efficacy of interferon (IFN)-free therapy with asunaprevir and daclatasvir for patients infected with hepatitis C virus (HCV) genotype 1b. The characteristics of patients with an L31 or a Y93 variant in the nonstructural 5A region detected by a polymerase chain reaction invader assay were investigated.
Methods
In total, 201 patients with HCV genotype 1b were examined for L31F/M/V variants or a Y93H variant by the polymerase chain reaction invader assay.
Results
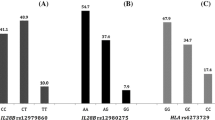
L31M and Y93H variants were detected in 4.6 and 21.4 % of patients, respectively. Patients with an L31M variant had no significant characteristics. Patients with a Y93H variant had significantly higher HCV RNA levels (6.5 ± 0.5 log copies per milliliter vs 6.1 ± 0.7 log copies per milliliter, p = 0.0002), higher frequency of mutant type of the IFN-sensitivity-determining region (88.4 % vs 71.7 %, p = 0.0251), and higher frequency of TT genotype at rs8099917 of IL28B (91.7 % vs 54.3 %, p < 0.0001) than those with Y93 wild-type strains. Multivariate analysis identified HCV RNA levels [odds ratio (OR) 3.72, 95 % confidence interval (CI) 1.71–8.06, p = 0.0009] and TT genotype at rs8099917 (OR 7.45, 95 % CI 2.11–26.4, p = 0.0018) as factors associated with the presence of a Y93H variant.
Conclusion
The presence of a Y93H variant was associated with higher HCV RNA levels and TT genotype at rs8099917 of IL28B. Thus, patients with a Y93H variant may be ideal candidates for IFN-based therapy rather than IFN-free therapy, although the high viral load of these patients may reduce the response rate of IFN-based therapy.
Similar content being viewed by others
References
Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–67.
Marcellin P, Asselah T, Boyer N. Fibrosis and disease progression in hepatitis C. Hepatology. 2002;36:S47–56.
Asselah T, Marcellin P. Optimal IFN-free therapy in treatment-naive patients with HCV genotype 1 infection. Liver Int. 2015;35(Suppl 1):56–64.
Suzuki F, Akuta N, Suzuki Y, et al. Rapid loss of hepatitis C virus genotype 1b from serum in patients receiving a triple treatment with telaprevir (MP-424), pegylated interferon and ribavirin for 12 weeks. Hepatol Res. 2009;39:1056–63.
Manns MP, Markova AA, Calle Serrano B, et al. Phase III results of boceprevir in treatment naive patients with chronic hepatitis C genotype 1. Liver Int. 2012;32(Suppl 1):27–31.
Hayashi N, Seto C, Kato M, et al. Once-daily simeprevir (TMC435) with peginterferon/ribavirin for treatment-naive hepatitis C genotype 1-infected patients in Japan: the DRAGON study. J Gastroenterol. 2014;49:138–47.
Hayashi N, Mobashery N, Izumi N. Vaniprevir plus peginterferon alfa-2a and ribavirin in treatment-experienced Japanese patients with hepatitis C virus genotype 1 infection: a randomized phase II study. J Gastroenterol. 2015;50:238–248.
Kumada H, Suzuki Y, Ikeda K, et al. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology. 2014;59:2083–91.
Afdhal N, Zeuzem S, Kwo P, et al. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med. 2014;370:1889–98.
Asselah T, Bruno S, Craxi A. HCV cirrhosis at the edge of decompensation: will paritaprevir with ritonavir, ombitasvir, dasabuvir, and ribavirin solve the need for treatment? J Hepatol. 2014;61:1430–3.
He Y, Staschke KA, Tan SL. HCV NS5A: a multifunctional regulator of cellular pathways and virus replication. In: Tan SL, editor. Hepatitis C viruses: genomes and molecular biology. Wymondham: Horizon Bioscience; 2006. p. 267–92.
Enomoto N, Sakuma I, Asahina Y, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81.
El-Shamy A, Nagano-Fujii M, Sasase N, et al. Sequence variation in hepatitis C virus nonstructural protein 5A predicts clinical outcome of pegylated interferon/ribavirin combination therapy. Hepatology. 2008;48:38–47.
Ohno O, Mizokami M, Wu RR, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201–7.
Tadokoro K, Suzuki F, Kobayashi M, et al. Rapid and high sensitive detection of Y93H amino acid substitution in HCV NS5A region using the PCR-Invader assay. Kanzo. 2014;55:720–2.
Nishikawa T, Hashimoto S, Kawabe N, et al. Factors correlating with acoustic radiation force impulse elastography in chronic hepatitis C. World J Gastroenterol. 2014;20:1289–97.
Akuta N, Suzuki F, Sezaki H, et al. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372–80.
Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–9.
Itakura J, Kurosaki M, Takada H, et al. Naturally occurring, resistance-associated hepatitis C virus NS5A variants are linked to IL28B genotype and are sensitive to interferon-based therapy. Hepatol Res. 2015. doi:10.1111/hepr.12474.
Uchida Y, Kouyama J, Naiki K, et al. A novel simple assay system to quantify the percent HCV-RNA levels of NS5A Y93H mutant strains and Y93 wild-type strains relative to the total HCV-RNA levels to determine the indication for antiviral therapy with NS5A inhibitors. PLoS One. 2014;9:e112647.
Miura M, Maekawa S, Sato M, et al. Deep sequencing analysis of variants resistant to the non-structural 5A inhibitor daclatasvir in patients with genotype 1b hepatitis C virus infection. Hepatol Res. 2014;44:E360–7.
Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401.
Shimada N, Toyoda H, Tsubota A, et al. Baseline factors and very early viral response (week 1) for predicting sustained virological response in telaprevir-based triple combination therapy for Japanese genotype 1b chronic hepatitis C patients: a multicenter study. J Gastroenterol. 2014;49:1485–94.
Chayama K, Hayes CN, Yoshioka K, et al. Factors predictive of sustained virological response following 72 weeks of combination therapy for genotype 1b hepatitis C. J Gastroenterol. 2011;46:545–55.
Taguchi T, Nagano-Fujii M, Akutsu M, et al. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol. 2004;85:959–69.
Choi YW, Tan YJ, Lim SG, et al. Proteomic approach identifies HSP27 as an interacting partner of the hepatitis C virus NS5A protein. Biochem Biophys Res Commun. 2004;318:514–9.
Shi ST, Polyak SJ, Tu H, et al. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198–210.
Cheng D, Zhang L, Yang G, et al. Hepatitis C virus NS5A drives a PTEN-PI3 K/Akt feedback loop to support cell survival. Liver Int. 2014. doi:10.1111/liv.12733.
Acknowledgments
This study was supported by MEXT-Supported Program for the Strategic Research Foundation at Private Universities of the Japanese government and by the Ministry of Health, Labour and Welfare of the Japanese government. The authors thank Makiko Shimazaki and Akie Tsuda from the Department of Liver, Biliary Tract, and Pancreas Diseases, Fujita Health University, for assisting with data collection and analysis.
Conflict of interest
Kentaro Yoshioka received a research grant from MSD KK, serves a consultant to Sanwa Kagaku Kenkyusho Co., Ltd., and received lecture fees from Bristol-Myers KK. Naoto Kawabe received a research grant from Bristol-Myers KK.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kan, T., Hashimoto, S., Kawabe, N. et al. The clinical features of patients with a Y93H variant of hepatitis C virus detected by a PCR invader assay. J Gastroenterol 51, 63–70 (2016). https://doi.org/10.1007/s00535-015-1080-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-015-1080-1