Abstract
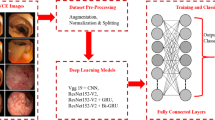
Ulcerative colitis (UC) can be classified as proctitis, left-sided colitis or pancolitis, usually with rectal involvement at the beginning. Mucosal carcinogenesis is one of the most severe complications of UC. Persistent inflammation of the rectal mucosa may be an essential cause of mucosal cancer, thus the detection of rectal inflammation is of great significance. In this paper, we propose a transfer learning model to classify enteroscopy images to achieve adequate detection of ulcerative proctitis. First, with the support of senior doctors, a dataset of ulcerative proctitis is created with 1450 endoscopic images. Then, with trained in the dataset, a new multi-model fusion network is proposed to classify ulcerative proctitis images. The proposed model combines three pre-trained models which are Xception, ResNet and DenseNet, and these pre-trained models are used to extract features from the images, then the extracted features are fed into a fully connected layer to predict the label of the input image. Experimental results show that, compared with other models, the proposed model has better performance, achieving the classification accuracy of 97.93%, the sensitivity of 99% and the specificity of 99%.








Similar content being viewed by others
References
De Chambrun, G.P., Tassy, B., Kollen, L., Dufour, G., Valats, J.C., Bismuth, M., Funakoshi, N., Panaro, F., Blanc, P.: Refractory ulcerative proctitis: How to treat it? Best Pract. Res. Clin. Gastroenterol. 32–33, 49–57 (2018)
Ordás, I., Eckmann, L., Talamini, M., Baumgart, D.C., Sandborn, W.J.: Ulcerative colitis. The Lancet 380(9853), 1606–1619 (2012)
Schroeder, K.W., Tremaine, W.J., Ilstrup, D.M.: Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. N. Engl. J. Med. 317(26), 1625–1629 (1987)
Ozawa, T., Ishihara, S., Fujishiro, M., Saito, H., Kumagai, Y., Shichijo, S., Aoyama, K., Tada, T.: Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest. Endosc. 89(2), 416–421 (2019)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521(7553), 436–444 (2015)
Esteva, A., Robicquet, A., Ramsundar, B., Kuleshov, V., DePristo, M., Chou, K., Cui, C., Corrado, G., Thrun, S., Dean, J.: A guide to deep learning in healthcare. Nat. Med. 25(1), 24–29 (2019)
Pan, S.J., Yang, Q.: A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22(10), 1345–1359 (2010)
Deng, J., Dong, W., Socher, R., Li, L., Kai, Li., Li, F.F.: Imagenet: a large-scale hierarchical image database. In: IEEE Conf. on Computer Vision and Pattern Recognition, pp. 248–255 (2009)
Esteva, A., Kuprel, B., Novoa, R.A., Ko, J., Swetter, S.M., Blau, H.M., Thrun, S.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115–118 (2017)
Amirreza, M., Gerald, S., Chunliang, W., Georg, D., Rupert, E., Isabella, E.: Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Meth. Prog. Biomed. 193, 105475 (2020)
Kooi, T., Litjens, G., Van Ginneken, B., Gubern-Mérida, A., Sánchez, C.I., Mann, R., Den Heeten, A., Karssemeijer, N.: Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 35, 303–312 (2017)
Pierrick, C., Boris, M., Michaël, C., Rémi, G., Baudouin, D.S., Vinh-Thong, T., Vincent, L., José, V.M.: AssemblyNet: a large ensemble of CNNs for 3D whole brain MRI segmentation. NeuroImage 219, 117026 (2020)
Wang, Y., Zu, C., Ma, Z., et al.: Patch-wise label propagation for MR brain segmentation based on multi-atlas images. Multimed. Syst. 25, 73–81 (2019)
Chatterjee, I., Kumar, V., Rana, B., et al.: Identification of changes in grey matter volume using an evolutionary approach: an MRI study of schizophrenia. Multimed. Syst. 26, 383–396 (2020)
De Fauw, J., Ledsam, J.R., Romera-Paredes, B., Nikolov, S., Tomasev, N., Blackwell, S., Askham, H., Glorot, X., O’Donoghue, B., Visentin, D., van den Driessche, G., Lakshminarayanan, B., Meyer, C., Mackinder, F., Bouton, S., Ayoub, K., Chopra, R., King, D., Karthikesalingam, A.: Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 24(9), 1342–1350 (2018)
Mehta, N., Lee, C., Mendonça, L., Raza, K., Braun, P., Duker, J., Waheed, N., Lee, A.: Model-to-Data approach for deep learning in optical coherence tomography intraretinal fluid segmentation. JAMA Ophthalmol. 138, 1017–1024 (2020)
Xi, X., Meng, X., Yang, L., et al.: Automated segmentation of choroidal neovascularization in optical coherence tomography images using multi-scale convolutional neural networks with structure prior. Multimed. Syst. 25, 95–102 (2019)
Cireşan, D.C., Giusti, A., Gambardella, L.M., Schmidhuber, J.: Mitosis detection in breast cancer histology images with deep neural networks. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013, Springer, Berlin, Heidelberg, pp. 411–418 (2013)
Catherine, P.J., Yijiang, C., Andrew, R.J., Matthew, B.P., Clarissa, A.C., Miroslav, S., Jeffrey, B.H., Jarcy, Z., Stephen, M.H., John, O.T., Paula, T., John, R.S., Laura, B., Anant, M.: Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. (2020). https://doi.org/10.1016/j.kint.2020.07.044
Horie, Y., Yoshio, T., Aoyama, K., Yoshimizu, S., Horiuchi, Y., Ishiyama, A., Hirasawa, T., Tsuchida, T., Ozawa, T., Ishihara, S., Kumagai, Y., Fujishiro, M., Maetani, I., Fujisaki, J., Tada, T.: Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest. Endosc. 89(1), 25–32 (2019)
Shichijo, S., Nomura, S., Aoyama, K., Nishikawa, Y., Miura, M., Shinagawa, T., Takiyama, H., Tanimoto, T., Ishihara, S., Matsuo, K., Tada, T.: Application of convolutional neural networks in the diagnosis of helicobacter pylori infection based on endoscopic images. EBioMedicine 25, 106–111 (2017)
Nakashima, H., Kawahira, H., Kawachi, H., Sakaki, N.: Endoscopic three-categorical diagnosis of Helicobacter pylori infection using linked color imaging and deep learning: a single-center prospective study (with video). Gastric. Cancer 23, 1033–1040 (2020)
Hirasawa, T., Aoyama, K., Tanimoto, T., Ishihara, S., Shichijo, S., Ozawa, T., Ohnishi, T., Fujishiro, M., Matsuo, K., Fujisaki, J., Tada, T.: Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 21(4), 653–660 (2018)
Ueyama, H., Kato, Y., Akazawa, Y., Yatagai, N., Komori, H., Takeda, T., Matsumoto, K., Ueda, K., Matsumoto, K., Hojo, M., Yao, T., Nagahara, A., Tada, T.: Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J. Gastroen, Hepatol (2020). https://doi.org/10.1111/jgh.15190
Cogan, T., Cogan, M., Tamil, L.: Mapgi: Accurate identification of anatomical landmarks and diseased tissue in gastrointestinal tract using deep learning. Comput. Biol. Med. 111, 103351 (2019)
Bopanna, S., Ananthakrishnan, A.N., Kedia, S., Yajnik, V., Ahuja, V.: Risk of colorectal cancer in Asian patients with ulcerative colitis: a systematic review and meta-analysis. Lancet Gastroenterol. 2(4), 269–276 (2017)
Chollet, F.: Xception: deep learning with depthwise separable convolutions. In: 2017 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 1251–1258 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Identity mappings in deep residual networks. In: Computer Vision – ECCV 2016, Springer International Publishing, Cham, pp. 630–645 (2016)
Huang, G., Liu, Z., Van, Der Maaten, L., Weinberger K.Q.: Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2261–2269 (2017)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 2818–2826 (2016)
Abd Ghani, M.K., Mohammed, M.A., Arunkumar, N., Mostafa, S.A., Ibrahim, D.A., Abdullah, M.K., Jaber, M.M., Abdulhay, E., Ramirez-Gonzalez, G., Burhanuddin, M.A.: Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques. Neural Comput. Appl. 32(3), 625–638 (2020)
Zeng, F., Fang, G., Yao, L.: A deep neural network for identifying dna n4-methylcytosine sites. Front. Genet. 11, 209 (2020)
Lu, X., Wang, X., Ding, L., Li, J., Gao, Y., He, K.: frDriver: a functional region driver identification for protein sequence. IEEE/ACM Trans. Comput. Biol. Bioinform. (2020). https://doi.org/10.1109/TCBB.2020.3020096
Xinguo, L., Qian, X., Li, X., Miao, Q., Peng, Shaoliang: DMCM: a data-adaptive mutation clustering method to identify cancer-related mutation clusters. Bioinformatics 35(3), 389–397 (2019)
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zeng, F., Li, X., Deng, X. et al. An image classification model based on transfer learning for ulcerative proctitis . Multimedia Systems 27, 627–636 (2021). https://doi.org/10.1007/s00530-020-00722-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00530-020-00722-0