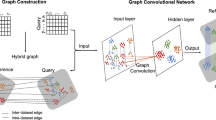
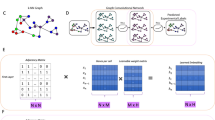
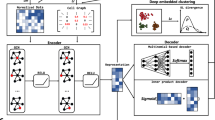
Abstract
Graph neural networks (GNNs) are reshaping our understanding of biomedicine and diseases by revealing the deep connections among genes and cells. As both algorithmic and biomedical technologies have advanced significantly, we are entering a transformative phase of personalized medicine. While pioneering tools like graph attention networks (GATs) and graph convolutional neural networks (Graph CNN) are advancing graph-based learning, the rise of single-cell sequencing techniques is reshaping our insights on cellular diversity and function. Numerous studies have combined GNNs with single-cell data, showing promising results. In this work, we highlight the GNN methodologies tailored for single-cell data over the recent years. We outline the diverse range of graph deep learning architectures that center on GAT methodologies. Furthermore, we underscore the several objectives of GNN strategies in single-cell data contexts, ranging from cell-type annotation, data integration and imputation, gene regulatory network reconstruction, clustering and many others. This review anticipates a future where GNNs become central to single-cell analysis efforts, particularly as vast omics datasets are continuously generated and the interconnectedness of cells and genes enhances our depth of knowledge in biomedicine.


Similar content being viewed by others
Data availability
No datasets were generated or analyzed during the current study.
References
Tang X, Huang Y, Lei J, Luo H, Zhu X (2019) The single-cell sequencing: new developments and medical applications. Cell Biosci 9:1–9
Rao A, Barkley D, França GS, Yanai I (2021) Exploring tissue architecture using spatial transcriptomics. Nature 596(7871):211–220
Wang R-S, Maron BA, Loscalzo J (2023) Multiomics network medicine approaches to precision medicine and therapeutics in cardiovascular diseases. Arterioscler Thromb Vasc Biol 43(4):493–503
Blencowe M, Arneson D, Ding J, Chen Y-W, Saleem Z, Yang X (2019) Network modeling of single-cell omics data: challenges, opportunities, and progresses. Emerg Topics Life Sci 3(4):379–398
Wu, L., Cui, P., Pei, J., Zhao, L., Song, L.: (2022) Graph neural networks. Springer, Singapore. pp. 27–37. https://doi.org/10.1007/978-981-16-6054-2_3
Georgousis S, Kenning MP, Xie X (2021) Graph deep learning: state of the art and challenges. IEEE Access 9:22106–22140
Ruiz L, Gama F, Ribeiro A (2020) Gated graph recurrent neural networks. IEEE Trans Signal Process 68:6303–6318
Kipf, T.N., Welling, M.: Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907 (2016)
Kipf, T.N., Welling, M.: Variational graph auto-encoders. arXiv preprint arXiv:1611.07308 (2016)
Mingshuo, N., Dongming, C., Dongqi, W.: Reinforcement learning on graph: A survey. arXiv e-prints, 2204 (2022)
Chen, L., Li, J., Peng, J., Xie, T., Cao, Z., Xu, K., He, X., Zheng, Z., Wu, B.: A survey of adversarial learning on graphs. arXiv preprint arXiv:2003.05730 (2020)
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., Bengio, Y.: Graph attention networks. arXiv preprint arXiv:1710.10903 (2017)
Labonne, M.: Hands-On Graph Neural Networks Using Python. Packt, ??? (2023)
Wang J, Ma A, Chang Y, Gong J, Jiang Y, Qi R, Wang C, Fu H, Ma Q, Xu D (2021) scgnn is a novel graph neural network framework for single-cell rna-seq analyses. Nature commun 12(1):1882
Shao X, Yang H, Zhuang X, Liao J, Yang P, Cheng J, Lu X, Chen H, Fan X (2021) scdeepsort: a pre-trained cell-type annotation method for single-cell transcriptomics using deep learning with a weighted graph neural network. Nucleic acids research 49(21):122–122
Hou W, Ji Z, Ji H, Hicks SC (2020) A systematic evaluation of single-cell rna-sequencing imputation methods. Genome biology 21:1–30
Rao, J., Zhou, X., Lu, Y., Zhao, H., Yang, Y.: Imputing single-cell rna-seq data by combining graph convolution and autoencoder neural networks. iscience (2021)
Feng X, Fang F, Long H, Zeng R, Yao Y (2022) Single-cell rna-seq data analysis using graph autoencoders and graph attention networks. Fronti Genet 13:1003711
Xu C, Cai L, Gao J (2021) An efficient scrna-seq dropout imputation method using graph attention network. BMC Bioinform 22:1–18
Feng X, Zhang H, Lin H, Long H (2023) Single-cell rna-seq data analysis based on directed graph neural network. Methods 211:48–60
Gu H, Cheng H, Ma A, Li Y, Wang J, Xu D, Ma Q (2022) scgnn 20: a graph neural network tool for imputation and clustering of single-cell rna-seq data. Bioinformatics 38(23):5322–5325
Wu X, Zhou Y (2022) Ge-impute: graph embedding-based imputation for single-cell rna-seq data. Brief Bioinform 23(5):313
Chen G, Liu Z-P (2022) Graph attention network for link prediction of gene regulations from single-cell rna-sequencing data. Bioinformatics 38(19):4522–4529
Buterez D, Bica I, Tariq I, Andrés-Terré H, Liò P (2022) Cellvgae: an unsupervised scrna-seq analysis workflow with graph attention networks. Bioinformatics 38(5):1277–1286
Baul S, Ahmed KT, Filipek J, Zhang W (2022) omicsgat: graph attention network for cancer subtype analyses. Int J Mol Sci 23(18):10220
Cheng Y, Ma X (2022) scgac: a graph attentional architecture for clustering single-cell rna-seq data. Bioinformatics 38(8):2187–2193
Huo Y, Guo Y, Wang J, Xue H, Feng Y, Chen W, Li X (2023) Integrating multi-modal information to detect spatial domains of spatial transcriptomics by graph attention network. J Genet Genom 50(9):720
Dong K, Zhang S (2022) Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. Nature Commun 13(1):1739
Abadi SAR, Laghaee SP, Koohi S (2023) An optimized graph-based structure for single-cell rna-seq cell-type classification based on non-linear dimension reduction. BMC Genom 24(1):1–13
Luo Z, Xu C, Zhang Z, Jin W (2021) A topology-preserving dimensionality reduction method for single-cell rna-seq data using graph autoencoder. Sci Rep 11(1):20028
Wang S, Zhang Y, Zhang Y, Wu W, Ye L, Li Y, Su J, Pang S (2023) scasgc: an adaptive simplified graph convolution model for clustering single-cell rna-seq data. Comput Biol Med 163:107152
Zhao J, Wang N, Wang H, Zheng C, Su Y (2021) Scdrha: a scrna-seq data dimensionality reduction algorithm based on hierarchical autoencoder. Front Genet 12:733906
So, E., Hayat, S., Kadambat Nair, S., Wang, B., Haibe-Kains, B.: (2023) Graphcomm: a graph-based deep learning method to predict cell-cell communication in single-cell rnaseq data. bioRxiv, 2023–04
Wang K, Li Z, You Z-H, Han P, Nie R (2023) Adversarial dense graph convolutional networks for single-cell classification. Bioinformatics 39(2):043
Shahir, J.A., Stanley, N., Purvis, J.E.: (2023) Cellograph: a semi-supervised approach to analyzing multi-condition single-cell rna-sequencing data using graph neural networks. bioRxiv, 2023–02
Ma A, Wang X, Li J, Wang C, Xiao T, Liu Y, Cheng H, Wang J, Li Y, Chang Y et al (2023) Single-cell biological network inference using a heterogeneous graph transformer. Nature Commun 14(1):964
Wang G, Zhao J, Yan Y, Wang Y, Wu AR, Yang C: (2023) Construction of a 3d whole organism spatial atlas by joint modeling of multiple slices. bioRxiv, 2023–02
Tang Z, Zhang T, Yang B, Su J, Song Q (2023) spaci: deciphering spatial cellular communications through adaptive graph model. Brief Bioinform 24(1):563
Dai X, Xu F, Wang S, Mundra PA, Zheng J (2021) Pike-r2p: Protein-protein interaction network-based knowledge embedding with graph neural network for single-cell rna to protein prediction. BMC Bioinform 22(6):1–16
Shan Y, Yang J, Li X, Zhong X, Chang Y (2023) Glae: A graph-learnable auto-encoder for single-cell rna-seq analysis. Inform Sci 621:88–103
Yu Z, Lu Y, Wang Y, Tang F, Wong K-C, Li X (2022) Zinb-based graph embedding autoencoder for single-cell rna-seq interpretations. Proc AAAI Conf Artif Intell 36:4671–4679
Shao X, Yang H, Zhuang X, Liao J, Yang Y, Yang P, Cheng J, Lu X, Chen H, Fan X: (2020) Reference-free cell-type annotation for single-cell transcriptomics using deep learning with a weighted graph neural network. BioRxiv 2020–05
Lee J, Kim S, Hyun D, Lee N, Kim Y, Park C (2023) Deep single-cell rna-seq data clustering with graph prototypical contrastive learning. Bioinformatics 39(6):342
Alghamdi N, Chang W, Dang P, Lu X, Wan C, Gampala S, Huang Z, Wang J, Ma Q, Zang Y et al (2021) A graph neural network model to estimate cell-wise metabolic flux using single-cell rna-seq data. Genome Res 31(10):1867–1884
Lin E, Liu B, Lac L, Fung D, Leung C, Hu P: (2023) scgmm-vgae: a gaussian mixture model-based variational graph autoencoder algorithm for clustering single-cell rna-seq data. Mach Learn Sci Technol
Bhadani R, Chen Z, An L (2023) Attention-based graph neural network for label propagation in single-cell omics. Genes 14(2):506
Yuan M, Chen L, Deng M (2022) scmra: a robust deep learning method to annotate scrna-seq data with multiple reference datasets. Bioinformatics 38(3):738–745
Yin Q, Liu Q, Fu Z, Zeng W, Zhang B, Zhang X, Jiang R, Lv H (2022) scgraph: a graph neural network-based approach to automatically identify cell types. Bioinformatics 38(11):2996–3003
Cao Z-J, Gao G (2022) Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nature Biotechnol 40(10):1458–1466
Li H, Sun Y, Hong H, Huang X, Tao H, Huang Q, Wang L, Xu K, Gan J, Chen H et al (2022) Inferring transcription factor regulatory networks from single-cell atac-seq data based on graph neural networks. Nature Mach Intell 4(4):389–400
Song Q, Su J, Zhang W (2021) scgcn is a graph convolutional networks algorithm for knowledge transfer in single cell omics. Nature Commun 12(1):3826
Liu Y, Zhang J, Wang S, Zhang W, Zeng X, Kwoh CK (2022) A heterogeneous graph cross-omics attention model for single-cell representation learning. In: 2022 IEEE international conference on bioinformatics and biomedicine (BIBM), pp 270–275. IEEE
Wu AP-Y, Singh R, Walsh CA, Berger B (2023) An econometric lens resolves cell-state parallax. bioRxiv, 2023
Yuan Y, Bar-Joseph Z (2020) Gcng: graph convolutional networks for inferring gene interaction from spatial transcriptomics data. Genome Biol 21(1):1–16
Li Y, Luo Y (2023) Spatial transcriptomic cell-type deconvolution using graph neural networks. bioRxiv
Cang Z, Ning X, Nie A, Xu M, Zhang J (2021) Scan-it: domain segmentation of spatial transcriptomics images by graph neural network. In: BMVC: proceedings of the british machine vision conference. british machine vision conference. vol 32. NIH Public Access
Kiselev VY, Andrews TS, Hemberg M (2019) Challenges in unsupervised clustering of single-cell rna-seq data. Nature Rev Genet 20(5):273–282
Heumos L, Schaar AC, Lance C, Litinetskaya A, Drost F, Zappia L, Lücken MD, Strobl DC, Henao J, Curion F et al (2023) Best practices for single-cell analysis across modalities. Nature Rev Genet 24(8):550
Maaten L, Hinton G (2008) Visualizing data using t-sne. J Mach Learn Res 9(11):104
McInnes L, Healy J, Melville J (2018) Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426
Lopez R, Regier J, Cole MB, Jordan MI, Yosef N (2018) Deep generative modeling for single-cell transcriptomics. Nature Methods 15(12):1053–1058
Risso D, Perraudeau F, Gribkova S, Dudoit S, Vert J-P (2018) A general and flexible method for signal extraction from single-cell rna-seq data. Nature Commun 9(1):284
Clarke ZA, Andrews TS, Atif J, Pouyabahar D, Innes BT, MacParland SA, Bader GD (2021) Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nature Protoc 16(6):2749–2764
Aalto A, Viitasaari L, Ilmonen P, Mombaerts L, Gonçalves J (2020) Gene regulatory network inference from sparsely sampled noisy data. Nature Commun 11(1):3493
Badia-i-Mompel P, Wessels L, Müller-Dott S, Trimbour R, Ramirez Flores RO, Argelaguet R, Saez-Rodriguez J (2023) Gene regulatory network inference in the era of single-cell multi-omics. Nature Rev Genet 1–16
Liu Z, Sun D, Wang C (2022) Evaluation of cell-cell interaction methods by integrating single-cell rna sequencing data with spatial information. Genome Biol 23(1):1–38
Xie Z, Li X, Mora A (2023) A comparison of cell-cell interaction prediction tools based on scrna-seq data. Biomolecules 13(8):1211
Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali T (2020) Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nature Methods 17(2):147–154
Tarashansky AJ, Xue Y, Li P, Quake SR, Wang B (2019) Self-assembling manifolds in single-cell rna sequencing data. Elife 8:48994
Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J et al (2017) Massively parallel digital transcriptional profiling of single cells. Nature Commun 8(1):14049
Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R et al (2018) Global characterization of t cells in non-small-cell lung cancer by single-cell sequencing. Nature Med 24(7):978–985
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161(5):1187–1201
Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM et al (2016) A single-cell transcriptomic map of the human and mouse pancreas reveals inter-and intra-cell population structure. Cell Syst 3(4):346–360
Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, Chen H, Wang J, Tang H, Ge W et al (2020) Construction of a human cell landscape at single-cell level. Nature 581(7808):303–309
Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F et al (2018) Mapping the mouse cell atlas by microwell-seq. Cell 172(5):1091–1107
Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini JL, Tran MN, Besich Z, Tippani M et al (2021) Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nature Neurosci 24(3):425–436
Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, Dang C (2012) Allen brain atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 41(D1):996–1008
Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F (2021) Highly sensitive spatial transcriptomics at near-cellular resolution with slide-seqv2. Nature Biotechnol 39(3):313–319
He S, Bhatt R, Brown C, Brown EA, Buhr DL, Chantranuvatana K, Danaher P, Dunaway D, Garrison RG, Geiss G et al (2022) High-plex imaging of rna and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nature Biotechnol 40(12):1794–1806
Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J et al (2018) Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361(6400):5691
Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C et al (2018) Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362(6416):5324
13 B.W.H..H.M.S.C.L...P.P.J..K.R., Medicine Creighton Chad J. 22 23 Donehower Lawrence A. 22 23 24 25, G., Systems Biology Reynolds Sheila 31 Kreisberg Richard B. 31 Bernard Brady 31 Bressler Ryan 31 Erkkila Timo 32 Lin Jake 31 Thorsson Vesteinn 31 Zhang Wei 33 Shmulevich Ilya 31, I., et al.: Comprehensive molecular portraits of human breast tumours. Nature 490(7418), 61–70 (2012)
Network CGAR et al (2014) Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507(7492):315
Luecken MD, Burkhardt DB, Cannoodt R, Lance C, Agrawal A, Aliee H, Chen AT, Deconinck L, Detweiler AM, Granados AA et al (2021) A sandbox for prediction and integration of dna, rna, and proteins in single cells. In: 35th conference on neural information processing systems (NeurIPS 2021) track on datasets and benchmarks
Buus TB, Herrera A, Ivanova E, Mimitou E, Cheng A, Herati RS, Papagiannakopoulos T, Smibert P, Odum N, Koralov SB (2021) Improving oligo-conjugated antibody signal in multimodal single-cell analysis. Elife 10:61973
Cao J, O’Day DR, Pliner HA, Kingsley PD, Deng M, Daza RM, Zager MA, Aldinger KA, Blecher-Gonen R, Zhang F et al (2020) A human cell atlas of fetal gene expression. Science 370(6518):7721
Zhang Q, Yang LT, Chen Z, Li P (2018) A survey on deep learning for big data. Information Fusion 42:146–157
Longo SK, Guo MG, Ji AL, Khavari PA (2021) Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nature Rev Genet 22(10):627–644
Brody, S., Alon, U., Yahav, E.: How attentive are graph attention networks? arXiv preprint arXiv:2105.14491 (2021)
Joshi-Tope, G., Gillespie, M., Vastrik, I., D’Eustachio, P., Schmidt, E., Bono, B., Jassal, B., Gopinath, G., Wu, G., Matthews, L., et al.: Reactome: a knowledgebase of biological pathways. Nucleic acids research 33(suppl_1), 428–432 (2005)
Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, Saez-Rodriguez J (2019) Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 29(8):1363–1375
Han H, Cho J-W, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E et al (2018) Trrust v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 46(D1):380–386
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lazaros, K., Koumadorakis, D.E., Vlamos, P. et al. Graph neural network approaches for single-cell data: a recent overview. Neural Comput & Applic (2024). https://doi.org/10.1007/s00521-024-09662-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00521-024-09662-6