Abstract
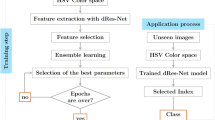
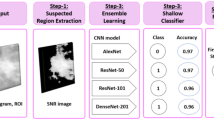
Histopathological diagnosis is the mainstay of present-day preventive medical care service to guide the therapy and treatment of breast cancer at an early stage. Manual examination of histologic data based on clinicians’ subjective knowledge is a time-consuming, labour-intensive, and costly method that necessitates clinical intervention and competence for a fair decision. In the recent work, we have developed an ensemble of five deep CNNs to classify three grades of breast cancer using quantitative image-based assessment of digital pathology slides without any manual intervention. To produce final predictions on the dataset, a fuzzy ranking algorithm is used. On the Databiox dataset, the suggested model attained an accuracy of 79%, 75%, 89%, and 82% at 4×, 10×, 20×, and 40× magnification, respectively. Furthermore, it has been observed that the stain-normalization strategy improves the model’s classification performance on the histopathological images. In this case, the Macenko stain-normalization technique is employed which further enhances the performance of the proposed ensemble model up to 80%, 100%, 100%, and 82% at 4×, 10×, 20×, and 40× magnification, respectively. Additionally, a comparative analysis with the existing state-of-the-art technique demonstrated the superiority of the proposed scheme.

Reproduced from https://pathology.jhu.edu/breast/staging-grade/















Similar content being viewed by others
Data availability
The dataset is publicly available at http://databiox.com.
References
Leong AS, Zhuang Z (2011) The changing role of pathology in breast cancer diagnosis and treatment. Pathobiology 78:99–114
Sravan M, Shankar M (2015) A current view on new cancer drugs (2014-USFDA approved) over old drugs. Int J Novel Trends Pharm Sci 5:198–208
Young RJ, Brown NJ, Reed MW, Hughes D, Woll PJ (2010) Angiosarcoma. Lancet Oncol 11:983–991
Jawad MA, Khursheed F (2022) Deep and dense convolutional neural network for multi category classification of magnification specific and magnification independent breast cancer histopathological images. Biomed Signal Process Control 78:103935
Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B (2009) Histopathological image analysis: a review. IEEE Rev Biomed Eng 2:147–171
Tellez D, Balkenhol M, Otte-Höller I, van de Loo R, Vogels R, Bult P, Wauters C, Vreuls W, Mol S, Karssemeijer N, Litjens G (2018) Whole-slide mitosis detection in H&E breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans Med Imaging 37:2126–2136
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
Oluogun WA, Adedokun KA, Oyenike MA, Adeyeba OA (2019) Histological classification, grading, staging, and prognostic indexing of female breast cancer in an African population: a 10-year retrospective study. Int J Health Sci 13:3
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19:403–410
He L, Long LR, Antani S, Thoma GR (2012) Histology image analysis for carcinoma detection and grading. Comput Methods Progr Biomed 107:538–556
Aksac A, Demetrick DJ, Ozyer T, Alhajj R (2019) BreCaHAD: a dataset for breast cancer histopathological annotation and diagnosis. BMC Res Notes 12:1–3
Kwon AY, Park HY, Hyeon J, Nam SJ, Kim SW, Lee JE, Yu JH, Lee SK, Cho SY, Cho EY (2019) Practical approaches to automated digital image analysis of Ki-67 labeling index in 997 breast carcinomas and causes of discordance with visual assessment. PLoS ONE 14:e0212309
Lloyd MC, Allam-Nandyala P, Purohit CN, Burke N, Coppola D, Bui MM (2010) Using image analysis as a tool for assessment of prognostic and predictive biomarkers for breast cancer: How reliable is it? J Pathol Inform 1:29
Balusamy B, Chilamkurti N, Beena LA, Poongodi T (2021) Blockchain and machine learning for e-healthcare systems, pp 1–481
Sharma S, Kumar S (2022) The Xception model: a potential feature extractor in breast cancer histology images classification. ICT Express 8(1):101–108
Kumaraswamy E, Kumar S, Sharma M (2023) An invasive ductal carcinomas breast cancer grade classification using an ensemble of convolutional neural networks. Diagnostics. pp 1–17. https://doi.org/10.3390/diagnostics13111977
Kumaraswamy E, Sharma S, Kumar S (2021) Invasive Ductal Carcinoma Grade Classification in Histopathological Images using Transfer Learning approach. In: 2021 IEEE Bombay Section Signature Conference (IBSSC), Gwalior, India, pp 1–6. https://doi.org/10.1109/IBSSC53889.2021.9673156
Hamida AB, Devanne M, Weber J, Truntzer C, Derangère V, Ghiringhelli F, Forestier G, Wemmert C (2021) Deep learning for colon cancer histopathological images analysis. Comput Biol Med 136:104730
Chouhan N, Khan A, Shah JZ, Hussnain M, Khan MW (2021) Deep convolutional neural network and emotional learning based breast cancer detection using digital mammography. Comput Biol Med 132:104318
Kawahara D, Tsuneda M, Ozawa S, Okamoto H, Nakamura M, Nishio T, Saito A, Nagata Y (2022) Stepwise deep neural network (stepwise-net) for head and neck auto-segmentation on CT images. Comput Biol Med 143:105295
Zavareh PH, Safayari A, Bolhasani H (2021) BCNet: a deep convolutional neural network for breast cancer grading
Aresta G, Araújo T, Kwok S, Chennamsetty SS, Safwan M, Alex V, Marami B, Prastawa M, Chan M, Donovan M (2019) Bach: grand challenge on breast cancer histology images. Med Image Anal 56:122–139
Chen H, Dou Q, Wang X, Qin J, Heng P (2016) Mitosis detection in breast cancer histology images via deep cascaded networks. In: Proceedings of the AAAI conference on artificial intelligence
Chen P-HC, Gadepalli K, MacDonald R, Liu Y, Kadowaki S, Nagpal K, Kohlberger T, Dean J, Corrado GS, Hipp JD (2019) An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat Med 25:1453–1457
Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson AN, Nelson HD, Pepe MS, Allison KH, Schnitt SJ (2015) Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313:1122–1132
George YM, Zayed HH, Roushdy MI, Elbagoury BM (2013) Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst J 8:949–964
Guo Z, Liu H, Ni H, Wang X, Su M, Guo W, Wang K, Jiang T, Qian Y (2019) A fast and refined cancer regions segmentation framework in whole-slide breast pathological images. Sci Rep 9:1–10
Kowal M, Filipczuk P, Obuchowicz A, Korbicz J, Monczak R (2013) Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput Biol Med 43:1563–1572
Marinelli RJ, Montgomery K, Liu CL, Shah NH, Prapong W, Nitzberg M, Zachariah ZK, Sherlock GJ, Natkunam Y, West RB (2007) The Stanford tissue microarray database. Nucleic Acids Res 36:D871–D877
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2015) A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng 63:1455–1462
Tao YK, Shen D, Sheikine Y, Ahsen OO, Wang HH, Schmolze DB, Johnson NB, Brooker JS, Cable AE, Connolly JL (2014) Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci 111:15304–15309
Bolhasani H, Amjadi E, Tabatabaeian M, Jassbi SJ (2020) A histopathological image dataset for grading breast invasive ductal carcinomas. Inform Med Unlocked 19:100341
Sharma S, Mehra R (2020) Conventional machine learning and deep learning approach for multi-classification of breast cancer histopathology images—a comparative insight. J Digit imaging 33:632–654
Sharma S, Mehra R (2020) Effect of layer-wise fine-tuning in magnification-dependent classification of breast cancer histopathological image. Vis Compute 36:1755–1769
Sharma S, Mehra R, Kumar S (2020) Optimised CNN in conjunction with efficient pooling strategy for the multi‐classification of breast cancer
Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, Schmitt C, Thomas NE (2009) A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE International symposium on biomedical imaging: from nano to macro. IEEE, pp 1107-1110
Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, Steiger K, Schlitter AM, Esposito I, Navab N (2016) Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging 35:962–1971
Huang J, Gong W, Chen H (2019) Menfish classification based on inception_V3 convolutional neural network. In: IOP Conference series: materials science and engineering. IOP Publishing, p 052099
Qin X, Wang Z (2019) Nasnet: a neuron attention stage-by-stage net for single image deraining
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258.
Chen H-Y, Su C-Y (2018) An enhanced hybrid MobileNet. In: 2018 9th International conference on awareness science and technology (iCAST). IEEE, pp 308–312.
Iandola F, Moskewicz M, Karayev S, Girshick R, Darrell T, Keutzer K (2014) Densenet: implementing efficient convnet descriptor pyramids
Targ S, Almeida D, Lyman (2016) Resnet in resnet: generalizing residual architectures
Naveenkumar M, Srithar S, Kumar BR, Alagumuthukrishnan S, Baskaran P (2021) InceptionResNetV2 for plant leaf disease classification. In: 2021 Fifth international conference on I-SMAC (IoT in social, mobile, analytics and cloud) (I-SMAC). IEEE, pp 1161–1167
Guan Q, Wang Y, Ping B, Li D, Du J, Qin Y, Lu H, Wan X, Xiang J (2019) Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: a pilot study. J Cancer 10:4876
Eliasziw M, Donner A (1991) Application of the McNemar test to non-independent matched pair data. Stat Med 10:1981–1991
Smith JK, Brown AR, Davis LM (2022) Enhancing breast cancer grading on the Databiox dataset using CNN and data augmentation. J Med Image Anal 25(5):635–647
Johnson RS, Parker ML, Garcia EL (2021) Transfer learning with ResNet-50 for improved breast cancer grading on the Databiox dataset. Int J Comput Vis 38(2):187–201
Rodriguez PA, Martinez SM, Nguyen TH (2023) Leveraging inception-v3 for fine-grained breast cancer grading on the Databiox dataset. Pattern Recogn Lett 29(7):932–944
Acknowledgements
The authors are immensely thankful to Prof. (Dr.) Ajith Abraham, Pro Vice Chancellor, Bennett University, Greater Noida, U.P., India, for providing all the necessary facilities and support during the execution of the work. Also, Dr Sumit Kumar would like to thank Dr Ashok Mittal, Chancellor, Lovely Professional University, Phagwara, Punjab, India, for constant support throughout the work.
Funding
There is no funding source.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharma, S., Kumar, S., Sharma, M. et al. An ensemble of deep CNNs for automatic grading of breast cancer in digital pathology images. Neural Comput & Applic 36, 5673–5693 (2024). https://doi.org/10.1007/s00521-023-09368-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-09368-1