Abstract
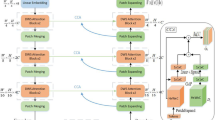
Computer-aided medical image segmentation has been applied widely in diagnosis and treatment to obtain clinically useful information of shapes and volumes of target organs and tissues. In the past several years, convolutional neural network (CNN)-based methods (e.g., U-Net) have dominated this area, but still suffered from inadequate long-range information capturing. Hence, recent work presented computer vision Transformer variants for medical image segmentation tasks and obtained promising performances. Such Transformers modeled long-range dependency by computing pair-wise patch relations. However, they incurred prohibitive computational costs, especially on 3D medical images (e.g., CT and MRI). In this paper, we propose a new method called Dilated Transformer, which conducts self-attention alternately in local and global scopes for pair-wise patch relations capturing. Inspired by dilated convolution kernels, we conduct the global self-attention in a dilated manner, enlarging receptive fields without increasing the patches involved and thus reducing computational costs. Based on this design of Dilated Transformer, we construct a U-shaped encoder–decoder hierarchical architecture called D-Former for 3D medical image segmentation. Experiments on the Synapse and ACDC datasets show that our D-Former model, trained from scratch, outperforms various competitive CNN-based or Transformer-based segmentation models at a low computational cost without time-consuming per-training process.





Similar content being viewed by others
Data availability statements
The data that support the findings of this study are openly available at https://doi.org/10.7303/syn3193805 and https://acdc.creatis.insa-lyon.fr/#challenge/5846c3366a3c7735e84b67ec.
References
Christ PF, Ettlinger F et al. (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. ArXiv:1702.05970
Pereira S, Pinto A (2016) Brain tumor segmentation using convolutional neural networks in MRI images. TMI 35(5):1240–1251
Brosch T, Tang LY, Yoo Y (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. TMI 35(5):1229–1239
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: CVPR. IEEE, pp 3431–3440
Korez R, Likar B, Pernuš F (2016) Model-based segmentation of vertebral bodies from MR images with 3D CNNs. In: MICCAI. Springer, pp 433–441
Zhou X, Ito T, Takayama R (2016) Three-dimensional CT image segmentation by combining 2D fully convolutional network with 3D majority voting. In: Deep learning and data labeling for medical applications. Springer, pp 111–120
Moeskops P, Wolterink JM (2016) Deep learning for multi-task medical image segmentation in multiple modalities. In: MICCAI. Springer, pp 478–486
Shakeri M, Tsogkas S, Ferrante E (2016) Sub-cortical brain structure segmentation using F-CNN’s. In: International symposium on biomedical imaging. IEEE, pp 269–272
Alansary A, Kamnitsas K, Davidson A (2016) Fast fully automatic segmentation of the human placenta from motion corrupted MRI. In: MICCAI. Springer, pp 589–597
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. In: MICCAI, pp 234–241
Wang C, MacGillivray T, Macnaught G et al (2018) A two-stage 3D Unet framework for multi-class segmentation on full resolution image. ArXiv:1804.04341
Çiçek, Ö, Abdulkadir A, Lienkamp SS (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: MICCAI. Springer, pp 424–432
Kamnitsas K, Ledig C, Newcombe VF (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. MIA 36:61–78
Drozdzal M, Vorontsov E, Chartrand G (2016) The importance of skip connections in biomedical image segmentation. In: Deep learning and data labeling for medical applications. Springer, pp 179–187
Ghafoorian M, Karssemeijer N, Heskes T (2016) Non-uniform patch sampling with deep convolutional neural networks for white matter hyperintensity segmentation. In: International symposium on biomedical imaging. IEEE, pp 1414–1417
Brosch T, Tang LY, Yoo Y (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. TMI 35(5):1229–1239
Milletari F, Navab N, Ahmadi S-A (2016) V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In: 3DV. IEEE, pp 565–571
Chen L-C, Papandreou G, Kokkinos I et al (2014) Semantic image segmentation with deep convolutional nets and fully connected CRFs. ArXiv:1412.7062
Chen L-C, Papandreou G, Kokkinos I (2017) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. TPAMI 40(4):834–848
Chen L-C, Papandreou G, Schroff F, et al (2017) Rethinking atrous convolution for semantic image segmentation. ArXiv:1706.05587
Chen L-C, Zhu Y, Papandreou G (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: ECCV, pp 801–818
Vaswani A, Shazeer N, Parmar N (2017) Attention is all you need. In: NIPS, vol 30
Devlin J, Chang M-W, Lee K, et al (2018) Bert: pre-training of deep bidirectional Transformers for language understanding. ArXiv:1810.04805
Dosovitskiy A, Beyer L, Kolesnikov A, et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. ArXiv:2010.11929
Touvron H, Cord M, Douze M (2021) Training data-efficient image transformers and distillation through attention. In: ICML. PMLR, pp 10347–10357
Carion N, Massa F, Synnaeve G (2020) End-to-end object detection with Transformers. In: ECCV. Springer, pp 213–229
Zhu X, Su W, Lu L, et al (2020) Deformable DETR: deformable transformers for end-to-end object detection. ArXiv:2010.04159
Wang X, Girshick R, Gupta A (2018) Non-local neural networks. In: CVPR. IEEE, pp 7794–7803
Liu Z, Lin Y, Cao Y, et al (2021) Swin transformers: hierarchical vision transformers using shifted windows. ArXiv:2103.14030
Wang W, Xie E, Li X, et al (2021) Pyramid vision transformers: a versatile backbone for dense prediction without convolutions. ArXiv:2102.12122
Zhang Z, Zhang H, Zhao L, et al (2021) Aggregating nested transformers. ArXiv:2105.12723
Zhou H-Y, Guo J, Zhang Y, et al (2021) nnFormer: interleaved transformers for volumetric segmentation. ArXiv:2109.03201
Sun Z, Cao S, Yang Y (2021) Rethinking transformer-based set prediction for object detection. In: ICCV, pp 3611–3620
Pan X, Xia Z, Song S (2021) 3D object detection with pointformer. In: CVPR. IEEE, pp 7463–7472
Yuan L, Chen Y, Wang T, et al (2021) Tokens-to-Token ViT: training vision Transformers from scratch on ImageNet. ArXiv:2101.11986
Yuan L, Hou Q, Jiang Z, et al (2021) VOLO: vision outlooker for visual recognition. ArXiv:2106.13112
Chen J, Lu Y, Yu Q, et al (2021) TransUNet: transformers make strong encoders for medical image segmentation. ArXiv:2102.04306
Hatamizadeh A, Tang Y, Nath V, et al (2021) UNETR: transformers for 3D medical image segmentation. ArXiv:2103.10504
Zhang Y, Liu H, Hu Q (2021) TransFuse: fusing transformers and CNNs for medical image segmentation. ArXiv:2102.08005
Xie Y, Zhang J, Shen C, et al (2021) CoTr: efficiently bridging CNN and transformer for 3D medical image segmentation. ArXiv:2103.03024
Cao H, Wang Y, Chen J, et al (2021) Swin-Unet: Unet-like pure Transformer for medical image segmentation. ArXiv:2105.05537
Lin A, Chen B, Xu J, et al (2021) DS-TransUNet: dual swin transformer U-Net for medical image segmentation. ArXiv:2106.06716
Huang X, Deng Z, Li D, et al (2021) MISSFormer: an effective medical image segmentation Transformer. ArXiv:2109.07162
El-Nouby A, Touvron H, Caron M, et al (2021) XCiT: cross-covariance image transformers. ArXiv:2106.09681
Wu Z, Liu Z, et al (2020) Lite Transformer with long-short range attention. ArXiv:2004.11886
Mehta S, Koncel-Kedziorski R, Rastegari M, Hajishirzi H (2020) DeFINE: DEep Factorized INput Token Embeddings for neural sequence modeling. ArXiv:1911.12385
Mehta S, Ghazvininejad M, Iyer S, et al (2020) DeLighT: very deep and light-weight transformer. CoRR
Shaw P, Uszkoreit J, Vaswani A (2018) Self-attention with relative position representations. ArXiv:1803.02155
Chu X, Tian Z, Zhang B, et al (2021) Conditional positional encodings for vision transformers. ArXiv:2102.10882
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: CVPR. IEEE, pp 1251–1258
Diakogiannis FI, Waldner F, Caccetta P (2020) ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. J Photogram Remote Sens 162:94–114
Ni Z-L, Bian G-B, Zhou X-H (2019) RAUNet: residual attention u-net for semantic segmentation of cataract surgical instruments. In: International conference on neural information processing. Springer, pp 139–149
Isensee F, Jaeger PF, Kohl SA (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18(2):203–211
Cai S, Tian Y, Lui H (2020) Dense-UNet: a novel multiphoton in vivo cellular image segmentation model based on a convolutional neural network. Quant Imaging Med Surg 10(6):1275
Zhou Z, Siddiquee MMR, Tajbakhsh N (2018) UNet++: a nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp 3–11
Huang H, Lin L, Tong R (2020) UNet 3+: a full-scale connected UNet for medical image segmentation. In: IEEE international conference on acoustics, speech and signal processing, pp 1055–1059
Peng C, Zhang X, Yu G (2017) Large kernel matters—improve semantic segmentation by global convolutional network. In: CVPR. IEEE, pp 4353–4361
Chen L-C, Papandreou G, Kokkinos I (2017) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. PAMI 40(4):834–848
Chen L-C, Zhu Y, Papandreou G (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: ECCV, pp 801–818
Roth HR, Shen C, Oda H (2018) A multi-scale pyramid of 3D fully convolutional networks for abdominal multi-organ segmentation. In: MICCAI, pp 417–425
Feng S, Zhao H, Shi F (2020) CPFNet: context pyramid fusion network for medical image segmentation. TMI 39(10):3008–3018
Heinrich MP, Oktay O, Bouteldja N (2019) OBELISK-Net: fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions. MIA 54:1–9
Li Z, Pan H, Zhu Y (2020) PGD-UNet: a position-guided deformable network for simultaneous segmentation of organs and tumors. In: International joint conference on neural networks. IEEE, pp 1–8
Han K, Xiao A, Wu E, et al (2021) Transformer in transformer. ArXiv:2103.00112
Zheng S, Lu J, Zhao H (2021) Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In: CVPR. IEEE, pp 6881–6890
Valanarasu JMJ, Oza P, et al (2021) Medical transformer: gated axial-attention for medical image segmentation. ArXiv:2102.10662
Çiçek Ö, Abdulkadir A, Lienkamp SS (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: MICCAI. Springer, pp 424–432
Ba JL, Kiros JR, Hinton GE (2016) Layer normalization. ArXiv:1607.06450
Kauderer-Abrams E (2017) Quantifying translation-invariance in convolutional neural networks. ArXiv:1801.01450
Wang W, Chen C, Ding M (2021) TransBTS: multimodal brain tumor segmentation using Transformer. In: MICCAI. Springer, pp 109–119
Xu G, Wu X, Zhang X, et al (2021) LeViT-UNet: make faster encoders with transformer for medical image segmentation. ArXiv:2107.08623
Deng J, Dong W, Socher R (2009) ImageNet: a large-scale hierarchical image database. In: CVPR. IEEE, pp 248–255
Bottou L (2012) Stochastic gradient descent tricks. In: Neural networks: tricks of the trade. Springer, pp 421–436
Mishra P, Sarawadekar K (2019) Polynomial learning rate policy with warm restart for deep neural network. In: IEEE region 10 conference, pp 2087–2092
Jadon S (2020) A survey of loss functions for semantic segmentation. In: IEEE conference on computational intelligence in bioinformatics and computational biology, pp 1–7
Yi-de M, Qing L, Zhi-Bai Q (2004) Automated image segmentation using improved PCNN model based on cross-entropy. In: International symposium on intelligent multimedia, video and speech processing, pp 743–746
Fu S, Lu Y, Wang Y (2020) Domain adaptive relational reasoning for 3D multi-organ segmentation. In: MICCAI. Springer, pp 656–666
Schlemper J, Oktay O, Schaap M (2019) Attention gated networks: learning to leverage salient regions in medical images. MIA 53:197–207
Dixon WJ, Mood AM (1946) The statistical sign test. J Am Stat Assoc 41(236):557–566
Hsu H, Lachenbruch PA (2014) Paired t test. Statistics Reference Online, Wiley StatsRef
Acknowledgements
This research was partially supported by the National Key R &D Program of China under Grant No. 2019YFB1404802, the National Natural Science Foundation of China under Grants No. 62176231 and 62106218, the Zhejiang public welfare technology research project under Grant No. LGF20F020013, and the Wenzhou Bureau of Science and Technology of China under Grant No. Y2020082. D. Z. Chen’s research was supported in part by NSF Grant CCF-1617735.
Funding
This research was partially supported by the National Key R &D Program of China under Grant No. 2019YFB1404802, the National Natural Science Foundation of China under Grants Nos. 62176231 and 62106218, the Zhejiang public welfare technology research project under Grant No. LGF20F020013 and the Wenzhou Bureau of Science and Technology of China under Grant No. Y2020082. D. Z. Chen’s research was supported in part by NSF Grant CCF-1617735.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Liao, K., Chen, J. et al. D-former: a U-shaped Dilated Transformer for 3D medical image segmentation. Neural Comput & Applic 35, 1931–1944 (2023). https://doi.org/10.1007/s00521-022-07859-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07859-1