Abstract
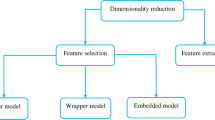
Feature selection (FS) is an important area of research in medicine and genetics. Cancer classification based on the microarray gene expression data is a challenge in this area due to its high-dimensional features and small sample size. This can negatively impact the performance of data mining and machine learning algorithms. FS is a key issue in reducing the size of the microarray, which is done to obtain useful information and eliminate redundant features. With the absence of a thorough investigation of the field, it is almost impossible for researchers to get an idea of how their work relates to existing studies and how it contributes to the research community. This paper provides a systematic mapping study to analyze and synthesize the studies conducted on the FS techniques in microarrays. To this end, 108 related articles published between 2000 and February 2022 were selected and reviewed based on five criteria: year and region, FS method adopted, dataset type, source of release, and type of evaluation software. Our main goal is to provide a fair idea to future researchers about the current situation of the field and future directions. The results of the study showed that classification is the most important task in FS. In a history-based evaluation, evolutionary methods were found to have the widest application to FS.












Similar content being viewed by others
References
Shah S, Kusiak A (2007) Cancer gene search with data-mining and genetic algorithms. Comput Biol Med 37:251–261. https://doi.org/10.1016/j.compbiomed.2006.01.007
Aminzadeh A, Ramzanpoor M, Molaarazi A, Kebria Ghasemi F, Roshandel G (2017) Relationship between rainfall and temperature with the incidence of cancer in Golestan Province, northern Iran. J Gorgan Univ Med Sci 19:80–85
Jain I, Jain VK, Jain R (2018) Correlation feature selection based improved-binary particle swarm optimization for gene selection and cancer classification. Appl Soft Comput 62:203–215. https://doi.org/10.1016/j.asoc.2017.09.038
Chlioui I, Idri A, Abnane I (2020) Data preprocessing in knowledge discovery in breast cancer: systematic mapping study. Comput Methods Biomech Biomed Eng Imaging Vis. https://doi.org/10.1080/21681163.2020.1730974
Idri A, Chlioui I, Ouassif BEl (2018) A systematic map of data analytics in breast cancer. In: Proceedings of the Australasian computer science week multiconference, proceedings of the Australasian computer science week multiconference. pp 1–10 https://doi.org/10.1145/3167918.3167930
Kadi I, Idri A, Fernandez-Aleman JL (2019) Systematic mapping study of data mining–based empirical studies in cardiology. Health Inform J 25:741–770. https://doi.org/10.1177/1460458217717636
Benhar H, Idri A, Fernandez-Aleman JL (2019) A systematic mapping study of data preparation in heart disease knowledge discovery. J Med Syst 43:1–17. https://doi.org/10.1007/s10916-018-1134-z
El Idrissi T, Idri A, Bakkoury Z (2018) Data mining techniques in diabetes self-management: A systematic map,. In: World conference on information systems and technologies. vol 162, pp 1142–1152. https://doi.org/10.1007/978-3-319-77712-2
Idri A, Benhar H, Fernandez-Aleman JL, Kadi I (2018) A systematic map of medical data preprocessing in knowledge discovery. Comput Methods Programs Biomed. https://doi.org/10.1016/j.cmpb.2018.05.007
Maldonado S, Weber R, Famili F (2014) Feature selection for high-dimensional class-imbalanced data sets using support vector machines. Inf Sci 286:228–246. https://doi.org/10.1016/j.ins.2014.07.015
Wang SL, Li X, Zhang S, Gui J, Huang DS (2010) Tumor classification by combining PNN classifier ensemble with neighborhood rough set based gene reduction. Comput Biol Med 40:179–189. https://doi.org/10.1016/j.compbiomed.2009.11.014
Duval B, Hao JK (2010) Advances in metaheuristics for gene selection and classification of microarray data. Brief Bioinform 11:127–141. https://doi.org/10.1093/bib/bbp035
AbdElNabi MLR, Wajeeh Jasim M, EL Bakry HM, Taha MHN, Khalifa NEM (2020) Breast and colon cancer classification from gene expression profiles using data mining techniques. Symmetry 12:1–16. https://doi.org/10.3390/sym12030408
Santhakumar D, Logeswari S (2020) Efficient attribute selection technique for leukaemia prediction using microarray gene data. Soft Comput 24:14265–14274. https://doi.org/10.1007/s00500-020-04793-z
Gumaei A, Sammouda R, Al-Rakhami M, AlSalman H, El-Zaart A (2021) Feature selection with ensemble learning for prostate cancer diagnosis from microarray gene expression. Health Inform J 27:1–13. https://doi.org/10.1177/1460458221989402
Fajila F, Yusof Y (2021) Incremental search for informative gene selection in cancer classification. Ann Emerg Technol Comput (AETiC) 5:15–21. https://doi.org/10.33166/AETiC.2021.02.002
Qasem SN, Saeed F (2021) Hybrid feature selection and ensemble learning methods for gene selection and cancer classification. Int J Adv Comput Sci Appl (IJACSA) 12:193–200. https://doi.org/10.14569/IJACSA.2021.0120225
Hamim M, Moudden El I, Hicham M, Hain M (2021) Gene selection for cancer classification: a new hybrid filter-C5.0 approach for breast cancer risk prediction. Adv Sci Technol Eng Syst J 6:871–878. https://doi.org/10.25046/aj060196
Chandrakar PK, Shrivas AK, Sahu N (2021) Design of a novel ensemble model of classification technique for gene-expression data of lung cancer with modified genetic algorithm. EAI Endorsed Trans Pervasive Health Technol 7:1–13. https://doi.org/10.4108/eai.8-1-2021.167845
Bolon-Canedo V, Sanchez-Marono N, Alonso-Betanzos A (2012) An ensemble of filters and classifiers for microarray data classification. Pattern Recogn 45:531–539. https://doi.org/10.1016/j.patcog.2011.06.006
Yu H, Ni J, Zhao J (2013) ACOSampling: an ant colony optimization-based undersampling method for classifying imbalanced DNA microarray data. Neurocomputing 101:309–318. https://doi.org/10.1016/j.neucom.2012.08.018
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif intell Res 16:321–357. https://doi.org/10.1613/jair.953
Liu B, Cui Q, Jiang T, Ma S (2004) A combinational feature selection and ensemble neural network method for classification of gene expression data. BMC Bioinform 5:1–12. https://doi.org/10.1186/1471-2105-5-136
Valentini G, Muselli M, Ruffino F (2004) Cancer recognition with bagged ensembles of support vector machines. Neurocomputing 56:461–466. https://doi.org/10.1016/j.neucom.2003.09.001
Yu Z, Chen H, You J, Liu J, Wong HS, Han Guoqiang, Li Le (2014) Adaptive fuzzy consensus clustering framework for clustering analysis of cancer data. IEEE/ACM Trans Comput Biol Bioinf 12:887–901. https://doi.org/10.1109/TCBB.2014.2359433
Sun L, Wang W, Xu J, Zhang S (2019) Improved LLE and neighborhood rough sets-based gene selection using Lebesgue measure for cancer classification on gene expression data. J Intell Fuzzy Syst 37:5731–5742. https://doi.org/10.3233/JIFS-181904
Potharaju SP, Sreedevi M (2019) Distributed feature selection (DFS) strategy for microarray gene expression data to improve the classification performance. Clin Epidemiol Glob Health 7:171–176. https://doi.org/10.1016/j.cegh.2018.04.001
Yu Z, Zhang Y, Chen CLP, You J, Wong HS, Dai D, Wu S, Zhang J (2018) Multiobjective semisupervised classifier ensemble. IEEE Trans Cybern 49:2280–2293. https://doi.org/10.1109/TCYB.2018.2824299
Zhao W, Wang G, Wang HB, Chen HL, Dong H, Zhao ZD (2011) A novel framework for gene selection. Int J Adv Comput Technol 3:184–191. https://doi.org/10.4156/ijact.vol3.issue3.18
Liu KH, Tong M, Xie ST, Yee Ng VT (2015) Genetic programming based ensemble system for microarray data classification. Comput Math Methods Med 2015:1–11. https://doi.org/10.1155/2015/193406
Chen Z, Li J, Wei L, Xu W, Shi Y (2011) Multiple-kernel SVM based multiple-task oriented data mining system for gene expression data analysis. Expert Syst Appl 38:12151–12159. https://doi.org/10.1016/j.eswa.2011.03.025
Han F, Sun W, Ling QH (2014) A novel strategy for gene selection of microarray data based on gene-to-class sensitivity information. PLoS ONE 9:888–896. https://doi.org/10.1016/j.neunet.2011.05.010
Nagpal A, Singh V (2019) Feature selection from high dimensional data based on iterative qualitative mutual information. J Intell Fuzzy Syst 36:5845–5856. https://doi.org/10.3233/JIFS-181665
Wu XY, Wu ZY, Kang Li (2008) Identification of differential gene expression for microarray data using recursive random forest. Chin Med J 121:2492–2496. https://doi.org/10.1097/00029330-200812020-00005
Wang H, Jing X, Niu B (2017) A discrete bacterial algorithm for feature selection in classification of microarray gene expression cancer data. Knowl-Based Syst 126:8–19. https://doi.org/10.1016/j.knosys.2017.04.004
Piao H (2011) DNA microarray data analysis using a correlational Bayesian network. J Med Imaging Health Inform 1:366–370. https://doi.org/10.1166/jmihi.2011.1044
Sathya M, Manju Priya S (2020) Modified Whale Optimization Algorithm For Feature Selection In Micro Array Cancer Dataset. Int J Sci Technol Res 9:549–556
Leung YY, Chang CQ, Hung YS (2012) An integrated approach for identifying wrongly labelled samples when performing classification in microarray data. PLoS ONE 7:1–10. https://doi.org/10.1371/journal.pone.0046700
Islam AK, Jeong S, Bari AT, Lim CG, Jeon SH (2015) MapReduce based parallel gene selection method. Appl Intell 42:147–156. https://doi.org/10.1007/s10489-014-0561-x
Tang J, Zhou S (2016) A new approach for feature selection from microarray data based on mutual information. IEEE/ACM Trans Comput Biol Bioinf 13:1004–1015. https://doi.org/10.1109/TCBB.2016.2515582
Bolon-Canedo V, Sanchez-Marono N, Alonso-Betanzos A (2014) Data classification using an ensemble of filters. Neurocomputing 135:13–20. https://doi.org/10.1016/j.neucom.2013.03.067
Lai CM, Yeh WC, Chang Chung-Yi (2016) Gene selection using information gain and improved simplified swarm optimization. Neurocomputing 218:331–338. https://doi.org/10.1016/j.neucom.2016.08.089
Khaire UM, Dhanalakshmi R (2020) Stability investigation of improved whale optimization algorithm in the process of feature selection. Int J Data Min Boinform. https://doi.org/10.1080/02564602.2020.1843554
Li J, Wang Fei (2016) Towards unsupervised gene selection: a matrix factorization framework. IEEE/ACM Trans Comput Biol Bioinf 14:514–521. https://doi.org/10.1109/TCBB.2016.2591545
Zhou X, Tuck DP (2007) MSVM-RFE: extensions of SVM-RFE for multiclass gene selection on DNA microarray data. Bioinformatics 23:1106–1114. https://doi.org/10.1093/bioinformatics/btm036
Fortino V, Kinaret P, Fyhrquist N, Alenius H, Greco D (2014) A robust and accurate method for feature selection and prioritization from multi-class OMICs data. PLoS ONE 9:1–9. https://doi.org/10.1371/journal.pone.0107801
Jansi Rani M, Devaraj D (2019) Two-stage hybrid gene selection using mutual information and genetic algorithm for cancer data classification. J Med Syst 43:1–11. https://doi.org/10.1007/s10916-019-1372-8
Yan C, Ma J, Luo H, Zhang G, Luo J (2019) A novel feature selection method for high-dimensional biomedical data based on an improved binary clonal flower pollination algorithm. Hum Hered 84:34–46. https://doi.org/10.1159/000501652
Baliarsingh SK, Vipsita S, Muhammad K, Bakshi S (2019) Analysis of high-dimensional biomedical data using an evolutionary multi-objective emperor penguin optimizer. Swarm Evol Comput 48:262–273. https://doi.org/10.1016/j.swevo.2019.04.010
Venkataramana L, Jacob SG, Ramadoss R, Saisuma D, Haritha D, Manoja K (2019) Improving classification accuracy of cancer types using parallel hybrid feature selection on microarray gene expression data. Genes Genomics 41:1301–1313. https://doi.org/10.1007/s13258-019-00859-x
Dif N, Elberrichi Z (2019) An enhanced recursive firefly algorithm for informative gene selection. Int J Swarm Intell Res (IJSIR) 10:21–33. https://doi.org/10.4018/IJSIR.2019040102
Mekour N, Hamou RM, Amine A (2019) Filter/wrapper methods for gene selection and classification of microarray dataset. J Softw Innov (IJSI) 7:65–80. https://doi.org/10.4018/IJSI.2019070104
Alomari OA, Khader AT, Al-Betar MA, Abualigah LM (2017) Gene selection for cancer classification by combining minimum redundancy maximum relevancy and bat-inspired algorithm. Int J Data Min Bioinform 19:32–51. https://doi.org/10.1504/IJDMB.2017.088538
Aziz R, Verma CK, Srivastava N (2017) A novel approach for dimension reduction of microarray. Comput Biol Chem 71:161–169. https://doi.org/10.1016/j.compbiolchem.2017.10.009
Annavarapu CS, Dara S, Banka H (2016) Cancer microarray data feature selection using multi-objective binary particle swarm optimization algorithm. EXCLI J 15:460–473. https://doi.org/10.17179/excli2016-481
Tran B, Xue B, Zhang M (2016) Genetic programming for feature construction and selection in classification on high-dimensional data. Memet Comput 8:3–15. https://doi.org/10.1007/s12293-015-0173-y
Chhabra G, Vashisht V, Ranjan J (2019) Improving accuracy for cancer classification with gene selection. Int J Innov Technol Explor Eng (IJITEE) 8:192–199
Mohamed NS, Zainudin S, Othman ZA (2017) Metaheuristic approach for an enhanced mRMR filter method for classification using drug response microarray data. Expert Syst Appl 90:224–231. https://doi.org/10.1016/j.eswa.2017.08.026
Salem H, Attiya G, El-Fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134. https://doi.org/10.1016/j.asoc.2016.11.026
Brahim AB, Limam M (2016) A hybrid feature selection method based on instance learning and cooperative subset search. Pattern Recogn Lett 69:28–34. https://doi.org/10.1016/j.patrec.2015.10.005
Bennet J, Ganaprakasam C, Kumar N (2015) A hybrid approach for gene selection and classification using support vector machine. Int Arab J Inf Technol (IAJIT) 12:695–700
Hatami N, Chira C (2013) Diverse accurate feature selection for microarray cancer diagnosis. Intell Data Anal 17:697–716. https://doi.org/10.3233/IDA-130601
Boucheham A, Batouche M, Meshoul S (2015) Robust hybrid wrapper/filter biomarker discovery from gene expression data based on generalised Island model. Int J Comput Biol Drug Des 8:251–274
Park CH, Kim SB (2015) Sequential random k-nearest neighbor feature selection for high-dimensional data. Expert Syst Appl 42:2336–2342. https://doi.org/10.1016/j.eswa.2014.10.044
Gonzalez F, Belanche LA (2013) Feature selection for microarray gene expression data using simulated annealing guided by the multivariate joint entropy. Computacion y Sistemas 18:275–293. https://doi.org/10.13053/cys-18-2-1473
Han F, Yang S, Guan J (2015) An effective hybrid approach of gene selection and classification for microarray data based on clustering and particle swarm optimisation. Int J Data Min Bioinform 13:103–121. https://doi.org/10.1504/ijdmb.2015.071515
Dessì N, Pes B, Cannas LM (2015) An evolutionary approach for balancing effectiveness and representation level in gene selection. J Inf Technol Res (JITR) 8:16–33. https://doi.org/10.4018/jitr.2015040102
Wang A, An N, Chen G, Li L, Alterovitz G (2015) Accelerating wrapper-based feature selection with K-nearest-neighbor. Knowl-Based Syst 83:81–91. https://doi.org/10.1016/j.knosys.2015.03.009
Qiu X, Qiu Y, Feng G, Li P (2015) A sparse fuzzy c-means algorithm based on sparse clustering framework. Neurocomputing 157:290–295. https://doi.org/10.1016/j.neucom.2015.01.003
Mavroeidis D, Marchiori E (2014) Feature selection for k-means clustering stability: theoretical analysis and an algorithm. Data Min Knowl Disc 28:918–960. https://doi.org/10.1007/s10618-013-0320-3
Li X, Gong X, Peng X, Peng S (2014) SSiCP: a new svm based recursive feature elimination algorithm for multiclass cancer classification. Int J Multimed Ubiquitous Eng 9:347–360. https://doi.org/10.14257/ijmue.2014.9.6.33
Park DK, Jung EY, Lee SH, Lim JS (2015) A composite gene selection for DNA microarray data analysis. Multimed Tools Appl 74:9031–9041. https://doi.org/10.1007/s11042-013-1583-9
Prasartvit T, Banharnsakun A, Kaewkamnerdpong B, Achalakul T (2013) Reducing bioinformatics data dimension with ABC-kNN. Neurocomputing 116:367–381. https://doi.org/10.1016/j.neucom.2012.01.045
Li Z, Yang A, Chen X, Zeng L, Cao T (2014) A composite method for feature selection of microarray data. J Comput Theor Nanosci 11:472–476. https://doi.org/10.1166/jctn.2014.3382
Sumathi A, Santhoshkumar S, Sakthivel NK (2012) Development of an efficient data mining classifier with microarray data set for gene selection and classification. J Theor Appl Inf Technol 35:208–214
Revathy N, Balasubramanian R (2012) GA-SVM wrapper approach for gene ranking and classification using expressions of very few genes. J Theor Appl Inf Technol 40:113–119
Porto-Diaz I, Bolon-Canedo V, Alonso-Betanzos A, Fontenla-Romero O (2011) A study of performance on microarray data sets for a classifier based on information theoretic learning. Neural Netw 24:888–896. https://doi.org/10.1016/j.neunet.2011.05.010
Du W, Sun Y, Wang Y, Cao Z, Zhang C, Liang Y (2013) A novel multi-stage feature selection method for microarray expression data analysis. Int J Data Min Bioinform 7:58–77. https://doi.org/10.1504/ijdmb.2013.050977
Jeyachidra J, Punithavalli M, Jeyachidra J (2015) A Novel Distinguishability Based Weighted Feature Selection Algorithms for Improved Classification of Gene Microarray. 11:443–452. https://doi.org/10.3844/jcssp.2015.443.452
Sungheetha A, Suganthi J (2013) An efficient clustering-classification method in an information gain NRGA-KNN algorithm for feature selection of micro array data. Life Sci J 10:691–700
Apiletti D, Baralis E, Bruno G, Fiori A (2012) Maskedpainter: feature selection for microarray data analysis. Intell Data Anal 16:717–737. https://doi.org/10.3233/IDA-2012-0546
Luo L, Ye L, Luo M, Huang D, Peng H, Yang F (2011) Methods of forward feature selection based on the aggregation of classifiers generated by single attribute. Comput Biol Med 41:435–441. https://doi.org/10.1016/j.compbiomed.2011.04.005
Mahmoodian H, Marhaban Hamiruce M, Abdulrahim R, Rosli R, Saripan I (2011) Using fuzzy association rule mining in cancer classification. Aust Phys Eng Sci Med 34:41–54. https://doi.org/10.1007/s13246-011-0054-8
Chuang LY, Ke CH, Chang HW, Yang CH (2009) A two-stage feature selection method for gene expression data. OMICS 13:127–137. https://doi.org/10.1089/omi.2008.0083
Chuang LY, Ke CH, Chang HW, Yang CH (2008) An evolutionary algorithm approach to optimal ensemble classifiers for DNA microarray data analysis. IEEE Trans Evol Comput 12:377–388. https://doi.org/10.1109/TEVC.2007.906660
Debnath R, Kurita T (2010) An evolutionary approach for gene selection and classification of microarray data based on SVM error-bound theories. Biosystems 100:39–46. https://doi.org/10.1016/j.biosystems.2009.12.006
Wang X, Gotoh O (2009) Accurate molecular classification of cancer using simple rules. BMC Med Genomics 2:1–23. https://doi.org/10.1186/1755-8794-2-64
Zhu S, Wang D, Yu K, Li T, Gong Y (2008) Feature selection for gene expression using model-based entropy. IEEE/ACM Trans Comput Biol Bioinf 7:25–36. https://doi.org/10.1109/TCBB.2008.35
Zhang LJ, Li ZJ, Chen HW (2008) Handling gene redundancy in microarray data using grey relational analysis. Int J Data Min Bioinform 2:134–144. https://doi.org/10.1504/IJDMB.2008.019094
Shen Q, Shi WM, Kong W (2008) Hybrid particle swarm optimization and tabu search approach for selecting genes for tumor classification using gene expression data. Comput Biol Chem 32:53–60. https://doi.org/10.1016/j.compbiolchem.2007.10.001
Kianmehr K, Zhang H, Nikolov K, Özyer T, Alhajj R (2007) Utilising neural network and support vector machine for gene expression classification. J Inf Knowl Manag 6:251–260. https://doi.org/10.1142/S0219649207001822
Chiang JH, Ho SH (2008) A combination of rough-based feature selection and RBF neural network for classification using gene expression data. IEEE Trans Nanobiosci 7:91–99. https://doi.org/10.1109/TNB.2008.2000142
Dash R, Misra B (2017) Gene selection and classification of microarray data: a Pareto DE approach. Intell Decis Technol 11:93–107. https://doi.org/10.3233/IDT-160280
Lin HY (2016) Gene discretization based on EM clustering and adaptive sequential forward gene selection for molecular classification. Appl Soft Comput 48:683–690. https://doi.org/10.1016/j.asoc.2016.07.015
Huang HL, Chang FL (2007) ESVM: evolutionary support vector machine for automatic feature selection and classification of microarray data. Biosystems 90:516–528. https://doi.org/10.1016/j.biosystems.2006.12.003
Yousef M, Jung S, Showe LC, Showe MK (2007) Recursive Cluster Elimination (RCE) for classification and feature selection from gene expression data. BMC Bioinform 8:1–12. https://doi.org/10.1186/1471-2105-8-144
Dashtban M, Balafar M, Suravajhala P (2018) Gene selection for tumor classification using a novel bio-inspired multi-objective approach. Genomics 110:10–17. https://doi.org/10.1016/j.ygeno.2017.07.010
Vanitha CDA, Devaraj D, Venkatesulu M (2015) Gene expression data classification using support vector machine and mutual information-based gene selection. Procedia Comput Sci 47:13–21. https://doi.org/10.1016/j.procs.2015.03.178
Nematzadeh H, Enayatifar R, Mahmud M, Akbari E (2019) Frequency based feature selection method using whale algorithm. Genomics 111:1946–1955. https://doi.org/10.1016/j.ygeno.2019.01.006
Bolon-Canedo V, Sanchez-Marono N, Alonso-Betanzos A (2015) Distributed feature selection: An application to microarray data classification. Appl Soft Comput 30:136–150. https://doi.org/10.1016/j.asoc.2015.01.035
Garro BA, Rodriguez K, Vazquez RA (2016) Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl Soft Comput 38:548–560. https://doi.org/10.1016/j.asoc.2015.10.002
Iam-On N, Boongoen T, Garrett S (2010) LCE: a link-based cluster ensemble method for improved gene expression data analysis. Bioinformatics 26:1513–1519. https://doi.org/10.1093/bioinformatics/btq226
Moayedikia A, Ong KL, Boo YL, Yeoh WGS, Jensen R (2017) Feature selection for high dimensional imbalanced class data using harmony search. Eng Appl Artif Intell 57:38–49. https://doi.org/10.1016/j.engappai.2016.10.008
Ram M, Najafi A, Shakeri MT (2017) Classification and biomarker genes selection for cancer gene expression data using random forest. Iran J Pathol 12:339–347. https://doi.org/10.30699/ijp.2017.27990
Liu KH, Zeng ZH, Ng VTY (2016) A hierarchical ensemble of ECOC for cancer classification based on multi-class microarray data. Inf Sci 349:102–118. https://doi.org/10.1016/j.ins.2016.02.028
Liu H, Liu L, Zhang H (2010) Ensemble gene selection for cancer classification. Pattern Recogn 43:2763–2772. https://doi.org/10.1016/j.patcog.2010.02.008
Balakrishnan K, Dhanalakshmi R, Khaire UM (2021) Improved salp swarm algorithm based on the levy flight for feature selection. J Supercomput 77:12399–12419. https://doi.org/10.1007/s11227-021-03773-w
Azadifar S, Ahmadi A (2021) A graph-based gene selection method for medical diagnosis problems using a many-objective PSO algorithm. BMC Med Inform Decis Mak 21:1–16. https://doi.org/10.1186/s12911-021-01696-3
Xie J, Wang M, Xu S, Huang Z, Grant PW (2021) The Unsupervised Feature Selection Algorithms Based on Standard Deviation and Cosine Similarity for Genomic Data Analysis. Front Genet 12:1–17. https://doi.org/10.3389/fgene.2021.684100
Zhang H (2021) Feature selection using approximate conditional entropy based on fuzzy information granule for gene expression data classification. Front Genet 12:1–8. https://doi.org/10.3389/fgene.2021.631505
Dash R (2021) An adaptive harmony search approach for gene selection and classification of high dimensional medical data. J King Saud Univ-Comput Inf Sci 33:195–207. https://doi.org/10.1016/j.jksuci.2018.02.013
Mahmood SG, Karyakos RS, Yacoob IM (2021) Hybrid gene selection method based on mutual information technique and dragonfly optimization algorithm. East-Eur J Enterp Technol 3:64–69. https://doi.org/10.15587/1729-4061.2021.233382
Sharifai AG, Zainol ZB (2021) Multiple filter-based rankers to guide hybrid grasshopper optimization algorithm and simulated annealing for feature selection with high dimensional multi-class imbalanced datasets. IEEE Access 9:74127–74142. https://doi.org/10.1109/ACCESS.2021.3081366
Hamim M, El Moudden I, Pant MD, Moutachaouik H, Hain M (2021) A hybrid gene selection strategy based on fisher and ant colony optimization algorithm for breast cancer classification. Int J Online Biomed Eng (iJOE) 17:148–163. https://doi.org/10.3991/ijoe.v17i02.19889
Baliarsingh SK, Vipsita S, Gandomi AH, Panda A, Bakshi S, Ramasubbareddy S (2020) Analysis of high-dimensional genomic data using MapReduce based probabilistic neural network. Comput Methods Programs Biomed 195:105–625. https://doi.org/10.1016/j.cmpb.2020.105625
Sharifai AG, Zainol Z (2020) The correlation-based redundancy multiple-filter approach for gene selection. Int J Data Min Bioinform 23:62–78. https://doi.org/10.1504/ijdmb.2020.10027155
Acknowledgements
This research was partially supported by Shokrolah Vahmiyan based on grant number (No.1/S/K). We thank Dr. Vahid Erfani-Moghaddam from Golestan University of Medical Sciences, who provided useful comments that greatly improved the research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix 1
Appendix 1
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vahmiyan, M., Kheirabadi, M. & Akbari, E. Feature selection methods in microarray gene expression data: a systematic mapping study. Neural Comput & Applic 34, 19675–19702 (2022). https://doi.org/10.1007/s00521-022-07661-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07661-z