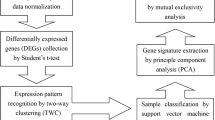
Abstract
Idiopathic pulmonary fibrosis (IPF) and nonspecific interstitial pneumonia (NSIP) are the two types of idiopathic interstitial pneumonia that are most prevalent. IPF and NSIP, often known as chronic interstitial pneumonia, must be differentiated from other forms of idiopathic interstitial pneumonia. However, distinguishing IPF from NSIP on radiographic imaging is challenging. Our goal in this work is to propose a novel approach to this clinical diagnostic challenge by distinguishing IPF from NSIP and healthy individuals via a complete systems biology analysis of existing microarray datasets. The Gene Expression Omnibus (GEO) database was searched, and two microarray datasets were identified. These datasets included normal, IPF, and NSIP samples. A second dataset was retrieved to validate further the built prediction models trained on the first dataset. Following the completion of the stages for data preparation and normalization, the profiles of gene expression were analyzed to determine the differentially expressed genes (DEGs). After that, we constructed module analysis and identified possible biomarkers by leveraging the prioritized and statistically significant DEGs to construct protein–protein interaction networks. The DEGs with the most important priority were also utilized to determine the implicated Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways and gene ontology (GO) enrichment analyses. Using the Kaplan–Meier approach, we performed three separate assessments of the gene biomarkers' effect on patients' chances of survival. In addition, the found genes were validated not just through several different categorization models, but also by analyzing the published experimental work on the target genes. A total of 32 distinct genes were found when comparing IPF to normal, NSIP to normal, and IPF to NSIP. This was accomplished by identifying seven (14 genes), six (7 genes), and eight (13 genes) modules, as well as three genes (i.e., C6, C5, STAT1). Results from GO analysis and the KEGG pathway evaluation showed evidence for biological processes, cellular components, and molecular activities. When considering the overall survival (OS), fast progression (FP), and post-progression survival (PPS) rates, the Kaplan–Meier analysis demonstrated that 27 out of 32, 16 out of 32, and 13 out of 32 genes were significant. Additionally, the identified biomarkers show high performance for the machine learning classification models. In addition, the scientific literature findings have validated each gene biomarker discovered for IPF, NSIP, and other lung-related conditions. The 32-mRNA signature shows promise as a gene set for IPF and NSIP and as a driver for treatments with the ability to predict and manage patients' survival rates accurately.








Similar content being viewed by others
Availability of data and material
Not applicable.
References
Adams J, Lawler J (1993) Extracellular matrix: the thrombospondin family. Curr Biol 3:188–190. https://doi.org/10.1016/0960-9822(93)90270-x
Åhrman E et al (2018) Quantitative proteomic characterization of the lung extracellular matrix in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. J Proteom 189:23–33. https://doi.org/10.1016/j.jprot.2018.02.027
American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS) (2000) Am J Respir Crit Care Med 161:646–664. https://doi.org/10.1164/ajrccm.161.2.ats3-00
Andersson-Sjöland A et al (2015) Versican in inflammation and tissue remodeling: the impact on lung disorders. Glycobiology 25:243–251. https://doi.org/10.1093/glycob/cwu120
Arias-Salgado EG et al (2019) Genetic analyses of aplastic anemia and idiopathic pulmonary fibrosis patients with short telomeres, possible implication of DNA-repair genes Orphanet J Rare Dis 14:82. https://doi.org/10.1186/s13023-019-1046-0
Augustine A, Robinson Vimala L, Irodi A, Mathew J, Pakkal M (2023) Diagnostic value of connective tissue disease related CT signs in usual interstitial pneumonia pattern of interstitial lung disease. Indian J Radiol Imaging 33:70–75. https://doi.org/10.1055/s-0042-1758876
Aviel-Ronen S et al (2008) Genomic markers for malignant progression in pulmonary adenocarcinoma with bronchioloalveolar features. Proc Natl Acad Sci USA 105:10155–10160. https://doi.org/10.1073/pnas.0709618105
Aye Y, Li M, Long MJC, Weiss RS (2015) Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 34:2011–2021. https://doi.org/10.1038/onc.2014.155
Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, Zhang L (2018) A potential risk factor of essential hypertension in case-control study: circular RNA hsa_circ_0037911. Biochem Biophys Res Commun 498:789–794. https://doi.org/10.1016/j.bbrc.2018.03.059
Bauer Y et al (2014) A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 52:217–231. https://doi.org/10.1165/rcmb.2013-0310OC
Beier CP et al (2005) FasL (CD95L/APO-1L) resistance of neurons mediated by phosphatidylinositol 3-kinase-Akt/protein kinase B-dependent expression of lifeguard/neuronal membrane protein 35. J Neurosci 25:6765–6774. https://doi.org/10.1523/jneurosci.1700-05.2005
Bennett D et al (2019) Calgranulin B and KL-6 in bronchoalveolar lavage of patients with IPF and NSIP. Inflammation 42:463–470. https://doi.org/10.1007/s10753-018-00955-2
Bornstein P, Sage EH (1994) Thrombospondins. Methods Enzymol 245:62–85. https://doi.org/10.1016/0076-6879(94)45006-4
Brereton CJ et al (2022) Pseudohypoxic HIF pathway activation dysregulates collagen structure-function in human lung fibrosis. Elife 11:e69348. https://doi.org/10.7554/eLife.69348
Bridges RS, Kass D, Loh K, Glackin C, Borczuk AC, Greenberg S (2009) Gene expression profiling of pulmonary fibrosis identifies Twist1 as an antiapoptotic molecular “rectifier” of growth factor signaling. Am J Pathol 175:2351–2361. https://doi.org/10.2353/ajpath.2009.080954
Brune JE, Chang MY, Altemeier WA, Frevert CW (2021) Type I interferon signaling increases versican expression and synthesis in lung stromal cells during influenza infection. J Histochem Cytochem 69:691–709. https://doi.org/10.1369/00221554211054447
Cabezas F, Farfán P, Marzolo MP (2019) Participation of the SMAD2/3 signalling pathway in the down regulation of megalin/LRP2 by transforming growth factor beta (TGF-ß1). PLoS ONE 14:e0213127. https://doi.org/10.1371/journal.pone.0213127
Carvalho B (2015) pd.hugene.1.0.st.v1: platform design info for affymetrix HuGene-1_0-st-v1. R package version 3141
Casanova N et al (2019) Low dose carbon monoxide exposure in idiopathic pulmonary fibrosis produces a CO signature comprised of oxidative phosphorylation genes. Sci Rep 9:14802. https://doi.org/10.1038/s41598-019-50585-3
Cedilak M et al (2019) Precision-cut lung slices from bleomycin treated animals as a model for testing potential therapies for idiopathic pulmonary fibrosis. Pulmon Pharmacol Ther 55:75–83. https://doi.org/10.1016/j.pupt.2019.02.005
Chandra A, Kahaleh B (2022) Systemic sclerosis (SSc) after COVID-19: a case report. Cureus 14
Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucl Acids Res 37:W305-311. https://doi.org/10.1093/nar/gkp427
Chen P, Cescon M, Bonaldo P (2013) Collagen VI in cancer and its biological mechanisms. Trends Mol Med 19:410–417. https://doi.org/10.1016/j.molmed.2013.04.001
Chen R (2013) Expression signature-guided functional genomics for gene discovery in non-small cell lung cancer. Stanford University
Chen W et al (2021) Anlotinib inhibits PFKFB3-driven glycolysis in myofibroblasts to reverse pulmonary fibrosis. Front Pharmacol 12:744826. https://doi.org/10.3389/fphar.2021.744826
Cho HK, Chung MP, Soo Lee K, Chung MJ, Han J, Kwon OJ, Yoo H (2022) Clinical characteristics and prognostic factors of fibrotic nonspecific interstitial pneumonia. Ther Adv Respir Dis. https://doi.org/10.1177/17534666221089468
Collado LM et al (2019) EP1. 03-04 analysis of post-surgical systemic inflammatory indexes after non-small cell lung cancer surgical intervention. J Thorac Oncol 14:S955
Confalonieri P et al (2022) Regeneration or repair? The role of alveolar epithelial cells in the pathogenesis of idiopathic pulmonary fibrosis (IPF). Cells. https://doi.org/10.3390/cells11132095
Coppieters F, Lefever S, Leroy BP, De Baere E (2010) CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat 31:1097–1108. https://doi.org/10.1002/humu.21337
Coussa RG et al (2013) WDR19: an ancient, retrograde, intraflagellar ciliary protein is mutated in autosomal recessive retinitis pigmentosa and in Senior-Loken syndrome. Clin Genet 84:150–159. https://doi.org/10.1111/cge.12196
Dalla-Torre CA et al (2006) Effects of THBS3, SPARC and SPP1 expression on biological behavior and survival in patients with osteosarcoma. BMC Cancer 6:237. https://doi.org/10.1186/1471-2407-6-237
Demšar J et al (2013a) Orange: data mining toolbox in Python. J Mach Learn Res 14:2349–2353
Demšar J et al (2013b) Orange: data mining toolbox in python. J Mach Learn Res 14:2349–2353
Dittel BN (2010) Depletion of specific cell populations by complement depletion. J vis Exp. https://doi.org/10.3791/1487
Elsayad AS, El-Desouky AI, Salem MM, Badawy M (2020) A deep learning H2O framework for emergency prediction in biomedical big data. IEEE Access 8:97231–97242
Emahazion T, Brookes AJ (1998) Mapping of the NDUFA2, NDUFA6, NDUFA7, NDUFB8, and NDUFS8 electron transport chain genes by intron based radiation hybrid mapping. Cytogenet Cell Genet 82:114. https://doi.org/10.1159/000015081
Glaspole I, Goh NS (2010) Differentiating between IPF and NSIP. Chronic Respir Dis 7:187–195. https://doi.org/10.1177/1479972310376205
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123:725–731. https://doi.org/10.1007/s10549-009-0674-9
Hinz B et al (2012) Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol 180:1340–1355. https://doi.org/10.1016/j.ajpath.2012.02.004
Ho CS, Noor SM, Nagoor NH (2018) MiR-378 and MiR-1827 regulate tumor invasion, migration and angiogenesis in human lung adenocarcinoma by targeting RBX1 and CRKL, respectively. J Cancer 9:331–345. https://doi.org/10.7150/jca.18188
Hossain MN, Sakemura R, Fujii M, Ayusawa D (2006) G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun 351:645–650. https://doi.org/10.1016/j.bbrc.2006.10.112
Hsu YL et al (2017) Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget 8:104831–104854. https://doi.org/10.18632/oncotarget.21022
Hu L, Smith TF, Goldberger G (2009) LFG: a candidate apoptosis regulatory gene family. Apoptosis 14:1255–1265. https://doi.org/10.1007/s10495-009-0402-2
Hu L, Yang Y, Tang Z, He Y, Luo X (2023) FCAN-MOPSO: an improved fuzzy-based graph clustering algorithm for complex networks with multi-objective particle swarm optimization. IEEE Trans Fuzzy Syst
Hu L, Zhang J, Pan X, Yan H, You Z-H (2021) HiSCF: leveraging higher-order structures for clustering analysis in biological networks. Bioinformatics 37:542–550
Hua X, Chen J, Wu L (2019) Identification of candidate biomarkers associated with apoptosis in melanosis coli: GNG5, LPAR3, MAPK8, and PSMC6. Biosci Rep 39
Huang DW, Sherman BT, Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res 37:1–13
Huang DW, Sherman BT, Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Huang WT, Yang X, He RQ, Ma J, Hu XH, Mo WJ, Chen G (2019) Overexpressed BSG related to the progression of lung adenocarcinoma with high-throughput data-mining, immunohistochemistry, in vitro validation and in silico investigation. Am J Transl Res 11:4835–4850
Johnston ID, Prescott RJ, Chalmers JC, Rudd RM (1997) British Thoracic Society study of cryptogenic fibrosing alveolitis: current presentation and initial management. Fibrosing Alveolitis Subcommittee of the Research Committee of the British Thoracic Society Thorax 52:38–44https://doi.org/10.1136/thx.52.1.38
Kakugawa T et al (2008) High serum concentrations of autoantibodies to HSP47 in nonspecific interstitial pneumonia compared with idiopathic pulmonary fibrosis. BMC Pulmon Med 8:23. https://doi.org/10.1186/1471-2466-8-23
Kang HC et al (2016) FAIM2, as a novel diagnostic maker and a potential therapeutic target for small-cell lung cancer and atypical carcinoid. Sci Rep 6:34022. https://doi.org/10.1038/srep34022
Karnitz LM, Zou L (2015) Molecular pathways: targeting ATR in cancer therapy. Clin Cancer Res 21:4780–4785. https://doi.org/10.1158/1078-0432.CCR-15-0479
Katzenstein AL, Fiorelli RF (1994) Nonspecific interstitial pneumonia/fibrosis. Histologic features and clinical significance. Am J Surg Pathol 18:136–147
Katzenstein AL, Myers JL, Mazur MT (1986) Acute interstitial pneumonia. A clinicopathologic, ultrastructural, and cell kinetic study. Am J Surg Pathol 10:256–267
Kehoe ER et al (2022) Biomarker selection and a prospective metabolite-based machine learning diagnostic for lyme disease. Sci Rep. https://doi.org/10.1038/s41598-022-05451-0
Khan FA et al (2016) CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget 7:52541–52552. https://doi.org/10.18632/oncotarget.9646
Khan MM et al (2021) An integrated multiomic and quantitative label-free microscopy-based approach to study pro-fibrotic signalling in <em>ex vivo</em> human precision-cut lung slices. Eur Respir J 58:2000221. https://doi.org/10.1183/13993003.00221-2020
Khaskhoussy R, Ayed YB (2022) Speech processing for early Parkinson’s disease diagnosis: machine learning and deep learning-based approach. Soc Netw Anal Min. https://doi.org/10.1007/s13278-022-00905-9
Kim HS, Lee MS (2007) STAT1 as a key modulator of cell death. Cell Signal 19:454–465. https://doi.org/10.1016/j.cellsig.2006.09.003
Kim JJ et al (2015) WSB1 promotes tumor metastasis by inducing pVHL degradation. Genes Dev 29:2244–2257. https://doi.org/10.1101/gad.268128.115
Koloko Ngassie M et al (2022a) Age-associated changes in the human lung extracellular matrix. In: D98. Targeting the scar: mechanisms and treatments for fibrotic lung disease. American Thoracic Society, p A5317
Koloko Ngassie ML et al (2022b) Age-associated differences in the human lung extracellular matrix.. bioRxiv:2022.2006.2016.496465. https://doi.org/10.1101/2022.06.16.496465
Kondoh Y et al (2005) Cyclophosphamide and low-dose prednisolone in idiopathic pulmonary fibrosis and fibrosing nonspecific interstitial pneumonia. Eur Respir J 25:528–533. https://doi.org/10.1183/09031936.05.00071004
Kyung SY et al (2014) Advanced glycation end-products and receptor for advanced glycation end-products expression in patients with idiopathic pulmonary fibrosis and NSIP. Int J Clin Exp Pathol 7:221–228
Lee J, Jang J, Park S-M, Yang S-R (2021) An update on the role of Nrf2 in respiratory disease: molecular mechanisms and therapeutic approaches. Int J Mol Sci. https://doi.org/10.3390/ijms22168406
Leng D, Yi J, Xiang M, Zhao H, Zhang Y (2020) Identification of common signatures in idiopathic pulmonary fibrosis and lung cancer using gene expression modeling. BMC Cancer 20:986. https://doi.org/10.1186/s12885-020-07494-w
Leslie KO (2005) Historical perspective: a pathologic approach to the classification of idiopathic interstitial pneumonias. Chest 128:513s–519s. https://doi.org/10.1378/chest.128.5_suppl_1.513S
Li S, You Z-H, Guo H, Luo X, Zhao Z-Q (2015) Inverse-free extreme learning machine with optimal information updating. IEEE Trans Cybern 46:1229–1241
Li T, Liu X, Yang A, Fu W, Yin F, Zeng X (2017a) Associations of tumor suppressor SPARCL1 with cancer progression and prognosis. Oncol Lett 14:2603–2610. https://doi.org/10.3892/ol.2017.6546
Li T, Liu X, Yang A, Fu W, Yin F, Zeng X (2017b) Associations of tumor suppressor SPARCL1 with cancer progression and prognosis (review). Oncol Lett 14:2603–2610. https://doi.org/10.3892/ol.2017.6546
Li Y, Guo L, Ying S, Feng G-H, Zhang Y (2020) Transcriptional repression of p21 by EIF1AX promotes the proliferation of breast cancer cells. Cell Prolifer 53:e12903. https://doi.org/10.1111/cpr.12903
Lin X, Wells DE, Kimberling WJ, Kumar S (1999) Human NDUFB9 gene: genomic organization and a possible candidate gene associated with deafness disorder mapped to chromosome 8q13. Hum Hered 49:75–80. https://doi.org/10.1159/000022848
Liu DW, Chen ST, Liu HP (2005) Choice of endogenous control for gene expression in nonsmall cell lung cancer. Eur Respir J 26:1002–1008. https://doi.org/10.1183/09031936.05.00050205
Liu H et al (2019) Prognostic and therapeutic potential of adenylate kinase 2 in lung adenocarcinoma. Sci Rep 9:17757. https://doi.org/10.1038/s41598-019-53594-4
Liu T, Gonzalez De Los Santos F, Hirsch M, Wu Z, Phan SH (2021) Noncanonical Wnt signaling promotes myofibroblast differentiation in pulmonary fibrosis. Am J Respir Cell Mol Biol 65:489–499https://doi.org/10.1165/rcmb.2020-0499OC
Llacua LA, Hoek A, de Haan BJ, de Vos P (2018) Collagen type VI interaction improves human islet survival in immunoisolating microcapsules for treatment of diabetes. Islets 10:60–68. https://doi.org/10.1080/19382014.2017.1420449
Lu P, Weaver VM, Werb Z (2012) The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196:395–406. https://doi.org/10.1083/jcb.201102147
Luo X, Liu Z, Jin L, Zhou Y, Zhou M (2021a) Symmetric nonnegative matrix factorization-based community detection models and their convergence analysis. IEEE Trans Neural Netw Learn Syst 33:1203–1215
Luo X, Qin W, Dong A, Sedraoui K, Zhou M (2020) Efficient and high-quality recommendations via momentum-incorporated parallel stochastic gradient descent-based learning. IEEE/CAA J Autom Sin 8:402–411
Luo X, Wu H, Wang Z, Wang J, Meng D (2021b) A novel approach to large-scale dynamically weighted directed network representation. IEEE Trans Pattern Anal Mach Intell 44:9756–9773
Maréchal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspect Biol. https://doi.org/10.1101/cshperspect.a012716
Molyneaux PL et al (2017) Host-microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 195:1640–1650. https://doi.org/10.1164/rccm.201607-1408OC
Mony JT, Schuchert MJ (2018) Prognostic implications of heterogeneity in intra-tumoral immune composition for recurrence in early stage lung cancer. Front Immunol 9:2298. https://doi.org/10.3389/fimmu.2018.02298
Myers JL, Katzenstein AL (2009) Beyond a consensus classification for idiopathic interstitial pneumonias: progress and controversies. Histopathology 54:90–103. https://doi.org/10.1111/j.1365-2559.2008.03173.x
Nair S, Bora-Singhal N, Perumal D, Chellappan S (2014) Nicotine-mediated invasion and migration of non-small cell lung carcinoma cells by modulating STMN3 and GSPT1 genes in an ID1-dependent manner. Mol Cancer 13:173. https://doi.org/10.1186/1476-4598-13-173
Nepusz T, Yu H, Paccanaro A (2012a) Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 9:471. https://doi.org/10.1038/nmeth.1938
https://www.nature.com/articles/nmeth.1938#supplementary-information
Nie MJ et al (2020) Clinical and prognostic significance of MYH11 in lung cancer. Oncol Lett 19:3899–3906. https://doi.org/10.3892/ol.2020.11478
Niemira M et al (2020) Molecular signature of subtypes of non-small-cell lung cancer by large-scale transcriptional profiling: identification of key modules and genes by weighted gene co-expression network analysis (WGCNA). Cancers. https://doi.org/10.3390/cancers12010037
Orchel J, Witek L, Kimsa M, Strzalka-Mrozik B, Kimsa M, Olejek A, Mazurek U (2012) Expression patterns of kinin-dependent genes in endometrial cancer. Int J Gynecol Cancer 22:937–944. https://doi.org/10.1097/IGC.0b013e318259d8da
Pardo A et al (2005) Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2:e251. https://doi.org/10.1371/journal.pmed.0020251
Parra ER, Araujo CA, Lombardi JG, Ab’Saber AM, Carvalho CR, Kairalla RA, Capelozzi VL (2012) Lymphatic fluctuation in the parenchymal remodeling stage of acute interstitial pneumonia, organizing pneumonia, nonspecific interstitial pneumonia and idiopathic pulmonary fibrosis. Braz J Med Biol Res 45:466–472. https://doi.org/10.1590/s0100-879x2012007500055
Patel AJ et al (2023) Lung cancer resection in patients with underlying usual interstitial pneumonia: a meta-analysis. BMJ Open Respir Res. https://doi.org/10.1136/bmjresp-2022-001529
Patnaik SR, Raghupathy RK, Zhang X, Mansfield D, Shu X (2015) The role of RPGR and its interacting proteins in ciliopathies. J Ophthalmol 2015:414781. https://doi.org/10.1155/2015/414781
Phan SH (2012) Genesis of the myofibroblast in lung injury and fibrosis. Proc Am Thorac Soc 9:148–152. https://doi.org/10.1513/pats.201201-011AW
Qi L et al (2019) MicroRNAs associated with lung squamous cell carcinoma: new prognostic biomarkers and therapeutic targets. J Cell Biochem. https://doi.org/10.1002/jcb.29216
Qiu X, Lin J, Liang B, Chen Y, Liu G, Zheng J (2021) Identification of hub genes and microRNAs associated with idiopathic pulmonary arterial hypertension by integrated bioinformatics analyses. Front Genet 12:667406. https://doi.org/10.3389/fgene.2021.636934
Raghu G et al (2011) An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 183:788–824. https://doi.org/10.1164/rccm.2009-040GL
Raghu G et al (2018) Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care Med 198:e44–e68. https://doi.org/10.1164/rccm.201807-1255ST
Raghu G et al (2015) An official ATS/ERS/JRS/ALAT clinical practice guideline: treatment of idiopathic pulmonary fibrosis. An update of the 2011 clinical practice guideline. Am J Respir Crit Care Med 192:e3–19. https://doi.org/10.1164/rccm.201506-1063ST
Rahmawati SF, Vos R, Bos IST, Kerstjens HAM, Kistemaker LEM, Gosens R (2022) Function-specific IL-17A and dexamethasone interactions in primary human airway epithelial cells. Sci Rep 12:11110. https://doi.org/10.1038/s41598-022-15393-2
Santos ARD et al (2021) Proteomic analysis of serum samples of paracoccidioidomycosis patients with severe pulmonary sequel. PLOS Negl Trop Dis 15:e0009714. https://doi.org/10.1371/journal.pntd.0009714
Schniering J et al (2020) Resolving phenotypic and prognostic differences in interstitial lung disease related to systemic sclerosis by computed tomography-based radiomics. medRxiv:2020.2006.2009.20124800. https://doi.org/10.1101/2020.06.09.20124800
Schoenfeld AR, Davidowitz EJ, Burk RD (2000) Elongin BC complex prevents degradation of von Hippel-Lindau tumor suppressor gene products. Proc Natl Acad Sci USA 97:8507–8512. https://doi.org/10.1073/pnas.97.15.8507
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
She K, Huang J, Zhou H, Huang T, Chen G, He J (2016) lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep 36:2673–2680. https://doi.org/10.3892/or.2016.5105
Shi K, Li N, Yang M, Li W (2019) Identification of key genes and pathways in female lung cancer patients who never smoked by a bioinformatics analysis. J Cancer 10:51–60. https://doi.org/10.7150/jca.26908
Shi X, He Q, Luo X, Bai Y, Shang M (2020) Large-scale and scalable latent factor analysis via distributed alternative stochastic gradient descent for recommender systems. IEEE Trans Big Data 8:420–431
Shu X et al (2012) Functional characterization of the human RPGR proximal promoter. Investig Ophthalmol vis Sci 53:3951–3958. https://doi.org/10.1167/iovs.11-8811
Shuai K, Schindler C, Prezioso VR, Darnell JE (1992) Activation of transcription by IFN-γ: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258:1808–1812. https://doi.org/10.1126/science.1281555
Stelzer G et al. (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform 54:1.30 (31-31.30.33)
Stepaniants S et al (2014) Genes related to emphysema are enriched for ubiquitination pathways. BMC Pulm Med 14:187. https://doi.org/10.1186/1471-2466-14-187
Sukumaran S, Jusko WJ, Dubois DC, Almon RR (2011) Light-dark oscillations in the lung transcriptome: implications for lung homeostasis, repair, metabolism, disease, and drug action. J Appl Physiol (Bethesda, Md: 1985) 110:1732–1747. https://doi.org/10.1152/japplphysiol.00079.2011
Sun W, Zhang L, Yan R, Yang Y, Meng X (2019) LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. OncoTargets Ther 12:3945–3954. https://doi.org/10.2147/ott.s196865
Sung A, Swigris J, Saleh A, Raoof S (2007) High-resolution chest tomography in idiopathic pulmonary fibrosis and nonspecific interstitial pneumonia: utility and challenges. Curr Opin Pulm Med 13:451–457. https://doi.org/10.1097/MCP.0b013e328273bc41
Szklarczyk D et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Tafti SF, Mokri B, Mohammadi F, Bakhshayesh-Karam M, Emami H, Masjedi MR (2008) Comparison of clinicoradiologic manifestation of nonspecific interstitial pneumonia and usual interstitial pneumonia/idiopathic pulmonary fibrosis: a report from NRITLD. Ann Thorac Med 3:140–145. https://doi.org/10.4103/1817-1737.43081
Tian Y, Tian X, Han X, Chen Y, Song C-Y, Jiang W-J, Tian D-L (2016a) ABCE1 plays an essential role in lung cancer progression and metastasis. Tumor Biol 37:8375–8382. https://doi.org/10.1007/s13277-015-4713-3
Tian Y, Tian X, Han X, Chen Y, Song CY, Jiang WJ, Tian DL (2016b) ABCE1 plays an essential role in lung cancer progression and metastasis. Tumour Biol 37:8375–8382. https://doi.org/10.1007/s13277-015-4713-3
Toledo LI, Murga M, Fernandez-Capetillo O (2011) Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol Oncol 5:368–373. https://doi.org/10.1016/j.molonc.2011.07.002
Travis WD, Matsui K, Moss J, Ferrans VJ (2000) Idiopathic nonspecific interstitial pneumonia: prognostic significance of cellular and fibrosing patterns: survival comparison with usual interstitial pneumonia and desquamative interstitial pneumonia. Am J Surg Pathol 24:19–33. https://doi.org/10.1097/00000478-200001000-00003
Uhlén M et al (2015) Tissue-based map of the human proteome. Science (New York, NY) 347:1260419
Uhlen M et al (2017) A pathology atlas of the human cancer transcriptome. Science (New York, NY) 357:eaan2507
Wang D, Han X, Li C, Bai W (2019) FBXL3 is regulated by miRNA-4735-3p and suppresses cell proliferation and migration in non-small cell lung cancer. Pathol Res Pract 215:358–365. https://doi.org/10.1016/j.prp.2018.12.008
Wang H, Yu L, Cui Y, Huang J (2022a) G protein subunit gamma 5 is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Dis Mark 2022:1313359. https://doi.org/10.1155/2022/1313359
Wang J et al (2022b) Characterizing cellular heterogeneity in fibrotic hypersensitivity pneumonitis by single-cell transcriptional analysis. Cell Death Discov 8:38. https://doi.org/10.1038/s41420-022-00831-x
Wang T et al (2015) XCR1 promotes cell growth and migration and is correlated with bone metastasis in non-small cell lung cancer. Biochem Biophys Res Commun 464:635–641. https://doi.org/10.1016/j.bbrc.2015.06.175
Wang X, Yang W, Yang Y, He Y, Zhang J, Wang L, Hu L (2022c) Ppisb: a novel network-based algorithm of predicting protein-protein interactions with mixed membership stochastic blockmodel. IEEE/ACM Trans Comput Biol Bioinform 20:1606–1612
Wang Y, Yella J, Chen J, McCormack FX, Madala SK, Jegga AG (2017) Unsupervised gene expression analyses identify IPF-severity correlated signatures, associated genes and biomarkers. BMC Pulm Med 17:133. https://doi.org/10.1186/s12890-017-0472-9
White ES, Lazar MH, Thannickal VJ (2003) Pathogenetic mechanisms in usual interstitial pneumonia/idiopathic pulmonary fibrosis. J Pathol 201:343–354. https://doi.org/10.1002/path.1446
Wijk SC et al (2022) Ciliated (FOXJ1(+)) cells display reduced ferritin light chain in the airways of idiopathic pulmonary fibrosis patients. Cells. https://doi.org/10.3390/cells11061031
Williams L, Layton T, Yang N, Feldmann M, Nanchahal J (2022) Collagen VI as a driver and disease biomarker in human fibrosis. FEBS J 289:3603–3629. https://doi.org/10.1111/febs.16039
Wu D, Luo X (2020) Robust latent factor analysis for precise representation of high-dimensional and sparse data. IEEE/CAA J Autom Sin 8:796–805
Xing R, Chen K-B, Xuan Y, Feng C, Xue M, Zeng Y-C (2016) RBX1 expression is an unfavorable prognostic factor in patients with non-small cell lung cancer. Surg Oncol 25:147–151. https://doi.org/10.1016/j.suronc.2016.05.006
Xu H et al (2016) Gene expression profiling analysis of lung adenocarcinoma. Braz J Med Biol. https://doi.org/10.1590/1414-431x20154861
Xue Z et al (2021) Combination therapy of tanshinone IIA and puerarin for pulmonary fibrosis via targeting IL6-JAK2-STAT3/STAT1 signaling pathways. Phytother Res 35:5883–5898. https://doi.org/10.1002/ptr.7253
Yamamoto M et al (2022) The differential diagnosis of IgG4-related disease based on machine learning. Arthritis Res Ther. https://doi.org/10.1186/s13075-022-02752-7
Yang Y, Li S, Cao J, Li Y, Hu H, Wu Z (2019) RRM2 regulated by LINC00667/miR-143-3p signal is responsible for non-small cell lung cancer cell progression oncotargets and therapy 12:9927–9939. https://doi.org/10.2147/ott.s221339
Ye J et al (2022) Biomarkers of connective tissue disease-associated interstitial lung disease in bronchoalveolar lavage fluid: a label-free mass spectrometry-based relative quantification study. J Clin Lab Anal 36:e24367. https://doi.org/10.1002/jcla.24367
Zhang W et al (2021) GNG5 is a novel oncogene associated with cell migration, proliferation, and poor prognosis in glioma. Cancer Cell Int 21:297. https://doi.org/10.1186/s12935-021-01935-7
Zhao B-W et al (2023) Fusing higher and lower-order biological information for drug repositioning via graph representation learning. IEEE Trans Emerg Top Comput
Zhao G et al (2019) Cullin5 deficiency promotes small-cell lung cancer metastasis by stabilizing integrin β1. J Clin Investig 129:972–987
Zheng X et al (2021) LDL receptor related protein 2 to promote colorectal cancer metastasis via enhancing GSK3β/β-catenin signaling. J Clin Oncol 39:e15507. https://doi.org/10.1200/JCO.2021.39.15_suppl.e15507
Zhu L, Li Q, Qi D, Niu F, Li Q, Yang H, Gao C (2019) Atherosclerosis-associated endothelial cell apoptosis by miRNA let7-b-mediated downregulation of HAS-2. J Cell Biochem. https://doi.org/10.1002/jcb.29537
Zhu L et al (2006) Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet 38:343–349. https://doi.org/10.1038/ng1721
Acknowledgements
The authors would like to thank the Research Office of Tabriz University of Medical Sciences for providing support under the support scheme (Grant no. 64907).
Funding
Funding was provided by Tabriz University of Medical Sciences (Grant no. 64907).
Author information
Authors and Affiliations
Contributions
BS—conceptualization, supervision, and project administration; EA and BS—data curation, methodology, and investigation; EA, SA, and SD—formal analysis; EA and BS—roles/writing—original draft; EA, BS, SA, and SD—writing—review and editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Amjad, E., Asnaashari, S., Dastmalchi, S. et al. An integrated study fusing systems biology and machine learning algorithms for genome-based discrimination of IPF and NSIP diseases: a new approach to the diagnostic challenge. Soft Comput 28, 5721–5749 (2024). https://doi.org/10.1007/s00500-023-09364-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-023-09364-6