Abstract
Key message
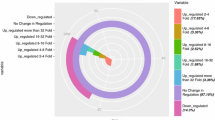
When Populus wutunensis seedlings are salt stressed, many genes involved in cellular metabolism, signal transduction, transport, osmotic regulation, and reactive oxygen species scavenging show elevated expression levels.
Abstract
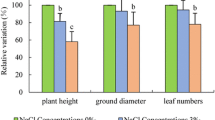
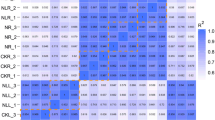
Populus wutunensis, a natural hybrid individual resulting from a cross between Populus × xiaozhuanica W. Y. Hsu et Liang, shows salt and drought tolerance and is fast-growing. It is widely cultivated in the Three-North Shelterbelt Forest area in China. To determine the transcriptome changes in P. wutunensis under salt stress, RNA-seq was conducted in salt-treated leaves and non-salt-treated control leaves. A total of 81,188 unigenes, with an average length of 982 bp, were obtained. In salt-treated leaves, 2466 differentially expressed genes were identified that differed from those in control leaves at any time point. Among them, 391 were upregulated and 44 were downregulated during all salt stress time points. They were mainly involved in cellular and metabolic processes, signal transduction, transport, osmotic regulation, and reactive oxygen species (ROS) scavenging. In addition, 2543 transcription factor genes were identified and divided into 60 families. qRT-PCR analysis revealed that the levels of differentially expressed genes were consistent with those from sequencing of the transcriptome under salt stress. These findings will increase our understanding of gene regulation in response to salt stress in P. wutunensis and are beneficial for future research on molecular-assisted breeding.








Similar content being viewed by others
Abbreviations
- ROS:
-
Reactive oxygen species
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- DEGs:
-
Differentially expressed genes
References
Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19(6):371–379
Dong J, Jiang YY, Chen RJ, Xu ZJ, Gao XL (2011) Isolation of a novel xyloglucan endotransgucosylase (OsXET9) gene from rice and analysis of the response of this gene to abiotic stresses. Afr J Biotechnol 10(76):17424–17434
Drerup MM, Schlücking K, Hashimoto K, Manishankar P, Steinhorst L, Kuchitsu K, Kudla J (2013) The cal-cineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHIJ. Mol Plant 6(2):559–569
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930
Golldack D, Luking I, Yang O (2011) Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 30(8):1383–1391
Guan PY, Wang J, Li H, Xie C, Zhang SZ, Wu CG, Yang GD, Yan K, Huang JG, Zheng CC (2018) Sensitive to SALT1, an endoplasmic reticulum-localized chaperone positively regulates salt resistance. Plant Physiol 178:1390–1405
Jia B, Wang W, Zhao Y, Lu X, Jiang G (2009) Growth of fast-growing Populus wutunensis with high saline-alkali tolerance. J Northeast For Univ 37(6):4–6
Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20:586–594
Li L, Kim BG, Cheong YH, PandeyGK LS (2006) Ca2+ signaling pathway regulated a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103:12625–12630
Li JB, Luan YS, Liu Z (2015) Overexpression of SpWRKY1promotes resistance to Phytophthoranicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol Plant 155(3):248–266
Li F, Zhang H, Zhao H, Gao T, Song A, Jiang J, Chen F, Chen S (2018) Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotech J16(7):1311–1321
Liu A, Xiao Z, Li MW, Wong FL, Yung WS, Ku YS, Wang Q, Wang X, Xie M, Yim AK, Chan TF, Lam HM (2019) Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ 42(1):98–114
Ma JL, Jiang GB, Yao SJ, Jin H (2013) Effects of apoplastic ion contents in the tender stems of two populus cultivars on gas exchange parameters under salt stress. Sci Silv 49(7):40–47
Nawar S, Buddenbaum H, Hill J (2015) Estimation of soil salinity using three quantitative methods based on visible and near-infrared reflectance spectroscopy: a casestudy from Egypt. Arab J Geosci 8(7):5127–5140
Sun T, Wang Y, Wang M, Li TT, Zhou Y, Wang XT, Wei SY, He GY, Yang GX (2015) Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol 15:260–269
Suzuki N, Koussevitzky S, Mittler R, Mittler G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35(2):259–270
Wang JC, Li BC, Meng YX, Ma XL, Lai Y, Si EJ, Ren PR, Shang XW, Wang JH (2015) Transcriptomic profiling of the salt-stress response in the halophyte Halogeton glomeratus. BMC genomics 16(1):169–183
Wang L, Du HY, Li TZ, Wuyun T (2018a) De novo transcriptome sequencing and identification of genes related to salt stress in Eucommiaulmoides oliver. Trees 32:151–163
Wang Z, Wang FX, Hong YC, Yao JJ, Ren ZZ, Shi HZ, Zhu JK (2018b) The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol Plant 11:1184–1197
Yan K, Shao H, Shao C, Chen P, Zhao SJ, Brestic M, Chen XB (2013) Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiologiae Plant 35(10):2867–2878
Yan YY, Wang SS, Wei M, Gong H (2018) Effect of different rootstocks on the salt stress tolerance in watermelon seedlings. Horticu Plant J 4(6):239–249
Yu CG, Xu S, Yin YL (2013) Transcriptome analysis of the Taxodium ‘Zhongshanshan 405’ roots in response to salinity stress. Plant Physiol Biochem 100:156–165
Zhao Y, Xing L, Wang X, Hou YJ, Gao J, Wang P, Duan CG, Zhu X, Zhu JK (2014) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7(328):ra53
Zhao Y, Zhang ZJ,Gao JH, Wang PC, Hu T, Wang ZG, Hou Y J, Wan YZ, Liu WS, XieSJ, Lu TJ, Xue L,Liu YJ,Macho AP, Tao WA, Bressan RA, Zhu JK (2018) Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep 23(11):3340–3351
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167(2):313–324
Acknowledgements
This work was supported by the Central Finance Forestry Science and Technology Promotion Demonstration Fund Project (Liao [2019] TG01) and the Fundamental Research Funds for the Central Universities(DC201501070103).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zou, J., Jin, H. Transcriptome sequencing and gene expression profiling of Populus wutunensis, a natural hybrid, during salinity stress. Trees 34, 1427–1438 (2020). https://doi.org/10.1007/s00468-020-02014-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-020-02014-6