Abstract
Key message
A strategy for effective utilization of RAPD marker data for sampling diverse entries was suggested and utilized for the development of mulberry core collection.
Abstract
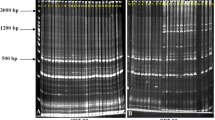
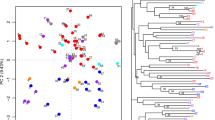
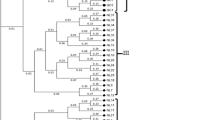
Mulberry (Morus spp.) is a perennial tree cultivated mainly for its foliage in sericulture industry and also known for its edible fruits, fodder, and valued timber. In recent years, mulberry cultivation is confronted with several abiotic and biotic stresses due to inimical climatic factors and this has necessitated the genetic improvement of the crop. Core collection is an efficient way of harnessing the trait variation and novel genes available in a natural gene pool for the development of improved elite lines. In this study, we analyzed 850 mulberry accessions assembled from 23 countries with separate sets of polymorphic RAPD markers along with a limited number of ISSR, SSR, and phenotypic markers. A total of 75 accessions were duplicated in 20 clusters among five natural groups. The limitations of the RAPD marker system like problem in cross gel comparison were tackled by adopting a novel “Groupwise sampling” approach. A mulberry core collection with 100 diverse entries was selected using maximization method implemented in MSTRAT software. The mean Dice dissimilarity coefficient computed from marker data was 0.308 among core entries. Indigenous and exotic entries were not discriminated in cluster and principal component analysis supporting the spread of mulberry far from the place of origin. Accessions belonging to two wild mulberry species from Andaman Islands and Himalayan region formed separate clusters indicating the geographical, reproductive, and taxonomic distinction. The identified core collection will be available for researchers for intensive mining of desirable alleles in mulberry improvement as well as in genome resequencing program.


Similar content being viewed by others
References
Balakrishna R, Sarakar A, Kumar JS (2002) Plasticity of some important traits in mulberry grown under shade and open condition. In: Dandin SB, Gupta VP (eds) Advances in Indian Sericulture Research, Proceedings of the national conference on strategies for sericulture research and development, Central Sericultural Research and Training Institute, Mysore, India, pp 4–7, Nov 16–18, 2000
Bouchez A, Causse M, Gallais A, Charcosset A (2002) Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics 162(4):1945–1959
Concibido V, la Vallee B, Mclaird P, Pineda N, Meyer J, Hummel L, Yang J, Wu K, Delannay X (2003) Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor Appl Genet 106(4):575–582
Das BC, Krishnaswami S (1965) Some observations on interspecific hybridization in mulberry. Indian J Sericult 4:1–8
Dulloo ME, Thormann I, Fiorino E, de Felice S, Rao VR, Snook L (2013) Trends in research using plant genetic resources from germplasm collections: from 1996 to 2006. Crop Sci 53(4):1217–1227
Franco J, Crossa J, Warburton ML, Taba S (2006) Sampling strategies for conserving maize diversity when forming core subsets using genetic markers. Crop Sci 42(2):854–864
Frankel OH (1984) Genetic perspectives of germplasm conservation. In: Arber W, Llimensee K, Peacock WJ, Starlinger P (eds) Genetic manipulation: impact on man and society. Cambridge University Press, Cambridge, pp 170–191
Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488(7412):535–539
Gouesnard B, Bataillon TM, Decoux G, Rozale C, Schoen DJ, David JL (2001) MSTRAT: an algorithm for building germplasm core collections by maximizing allelic or phenotypic richness. J Hered 92:93–94
Guruprasad (2011) Development of core collection of mulberry (Morus spp.) germplasm by molecular marker aided analysis. Ph.D. Thesis. University of Mysore
He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G et al. (2013) Draft genome sequence of the mulberry tree Morus notabilis. Nat Commun doi:10.1038/ncomms3445
Hu J, Zhu J, Xu HM (2000) Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor Appl Genet 101:264–268
Krishnan RR, Naik VG, Ramesh SR, Qadri SMH (2013) Microsatellite marker analysis reveals the events of the introduction and spread of cultivated mulberry in the Indian subcontinent. Plant Genet Resour. doi:10.1017/S1479262113000415
Le S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25(1):1–18
Meunier JR, Grimont PAD (1993) Factors affecting reproducibility of random amplified polymorphic DNA fingerprinting. Res Microbiol 144(5):373–379
Naik VG, Dandin SB (2005) Molecular characterization of some improved and promising mulberry varieties (Morus spp.) of India by RAPD and ISSR markers. Indian J Sericult 44(1):59–68
Naik VG, Dandin SB (2006) Identification of duplicate collections in the mulberry (Morus spp.) germplasm using RAPD analysis. Indian J Genet 66(4):287–292
Oakey HJ, Gibson LF, George AM (1998) Co-migration of RAPD-PCR amplicons from Aeromonas hydrophila. FEMS Microbiol Lett 164(1):35–38
Odong TL, Jansen J, van Eeuwijk FA, van Hintum TJL (2013) Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor Appl Genet 126(2):289–305
Paraguison RC, Faylon MPP, Flores EB, Cruz LC (2012) Improved RAPD-PCR for discriminating breeds of water buffalo. Biochem Genet 50(7):579–584
Penner GA, Bush A, Wise R, Kim W, Domier L, Kasha K et al (1993) Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories. Genome Res 2(4):341–345
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin. Accessed 14 March 2013
Ravindran S, Tikader A, Naik VG, Rao AA, Mukherjee P (1998) Distribution of mulberry species in India and its utilization. Indian J Plant Genet Resour 12(2):163–168
Sarkar A, Kumar JS, Datta RK (2000) Gradual improvement of mulberry varieties under irrigated condition in South India and optimal programme for varietal selection in the tropics. Sericologia 40:449–461
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327(5967):818–822
Thangavelu K, Mukherjee P, Tikader A, Ravindran S, Goel AK, Ananda Rao A, Naik VG, Shekar S (1997) Catalogue on mulberry (Morus spp.) germplasm, vol I. Central Sericultural Germplasm Resources Centre, Hosur
Upadhyaya HD, Ravishankar CR, Narasimhudu Y, Sarma NDRK, Singh SK, Varshney RK et al (2011) Identification of trait-specific germplasm and developing a mini core collection for efficient use of foxtail millet genetic resources in crop improvement. Field Crops Res 124(3):459–467
Vijayan K, Srivatsava PP, Nair CV, Awasthi AK, Tikader A, Sreenivasa B, Urs SR (2006) Molecular characterization and identification of markers associated with yield traits in mulberry using ISSR markers. Plant Breeding 125(3):298–301
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18(22):6531–6535
Zhang DP, Ghislain M, Huaman Z, Rodrguez F, Cervantes JC (1996) Identifying duplicates in sweetpotato germplasm using RAPD. International potato center program report 1995–1996. International Potato Center, Lima, pp 90–96
Acknowledgments
Authors thank the two anonymous reviewers for their critical comments and suggestions, which has helped us to improve the quality of the manuscript. VGN acknowledges the financial support received from the Central Silk Board, Ministry of Textiles, Government of India, Bangalore, India to undertake this research work in the project (Code #PIB-3370; Sanction Order No. CSB/RTI/PMCE/245(3)/2006-07 dated 24.07.2006). Guruprasad received Senior Research Fellowship under the project grant. The authors are grateful to the Director, Central Sericultural Germplasm Resource Center, Hosur, Tamil Nadu, India for providing mulberry germplasm leaf samples to carry out this work.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Alia.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guruprasad, Ramesh Krishnan, R., Dandin, S.B. et al. Groupwise sampling: a strategy to sample core entries from RAPD marker data with application to mulberry. Trees 28, 723–731 (2014). https://doi.org/10.1007/s00468-014-0984-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-014-0984-3