Abstract
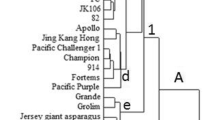
Phenotypic and genotypic variation and structure of 29 sour cherry (P. cerasus) and duke cherry (P. x gondouinii) genotypes from different regions of Iran were identified using random amplified polymorphic DNA (RAPD) markers and morphological characters. Furthermore, one Prunus mahaleb genotype was used as an outgroup for molecular analysis. For morphological analysis, 23 variables were recorded to detect similarities between and among studied sour and duke cherries. Most studied characteristics were showing a high degree of variability. Principal component analysis showed that the first three components explained a total of 73.87 % of the whole phenotypic variability. Based on the morphological cluster analysis, studied sour and duke cherry genotypes were placed into three main clusters. The first main cluster included 16 sour cherry genotypes. The second main cluster contained all duke cherry genotypes and eight sour cherry genotypes, while, only one sour cherry genotype was placed in third main cluster. For RAPD analysis, 17 primers generated a total of 233 discernible and reproducible bands across genotypes analyzed, out of which 214 (91.51 %) were polymorphic with varied band size from 300 to 3000 bp. According to the similarity matrix, the lowest similarity was obtained between P. mahaleb, as an outgroup, and sour cherry. Dendrogram based on molecular data separated genotypes according to their species and geographic origin. Low correlation was observed between the similarity matrices obtained based on morphological and RAPD data. The information obtained here could be valuable for devising strategies for conservation of Iranian sour and duke cherries.



Similar content being viewed by others
References
Antonius K, Aaltonen M, Uosukainen M, Hurme T (2012) Genotypic and phenotypic diversity in Finnish cultivated sour cherry (Prunus cerasus L.). Genet Resour Crop Evol 59:375–388
Badenes ML, Martinez-Calvo J, Llacer G (2000) Analysis of a germplasm collection of loquat (Eriobotrya japonica Lindl.). Euphytica 114:187–194
Beaver JA, Iezzoni AF (1993) Allozyme inheritance in tetraploid sour cherry (Prunus cerasus L.). J Amer Soc Hort Sci 118:873–877
Beaver JA, Iezzoni AF, Ramm CW (1995) Isozyme diversity in sour, sweet and ground cherry. Theor Appl Genet 90:847–852
Boritzki M, Plieske J, Struss D (2000) Cultivar identification in sweet cherry (Prunus avium L.) using AFLP and microsatellite markers. Acta Hort 538:505–510
Cai YL, Cao DW, Zhao GF (2007) Studies on genetic variation in cherry germplasm using RAPD analysis. Sci Hort 111:248–254
Cantini C, Cimato A, Sani G (1999) Morphological evaluation of olive germplasm present in Tuscany region. Euphytica 109:173–181
Cantini C, Iezzoni A, Lamboy AF, Boritzki M, Struss D (2001) DNA fingerprinting of tetraploid cherry germplasm using simple sequence repeats. J Am Soc Hort Sci 126:205–209
Corts RM, Rodrigues LC, Marcide JMO, Sanches RP (2008) Characterization of sour (Prunus cerasus L.) and sweet cherry (Prunus avium L.) varieties with five isozyme systems. Revista Brasileira de Fruticultura 30:154–158
Demirsoy LT, Demir H, Demirsoy A, Okumuş Y, Kaçar A (2008) Identification of some sweet cherry cultivars grown in Amasya by RAPD markers. Acta Hort 795:146–152
Dever MC, Macdonald RA, Chiff MA, Lane WD (1996) Sensory evaluation of sweet cherry cultivars. Hort Sci 31:150–153
Doyle JJ, Doyle JL (1987) A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
FAOSTAT (2010) At: http://faostat.fao.org/site/567/DesktopDefauH.aspx?PageID=567
Futuyma DJ (1986) Evolutionary biology, 2nd edn. Sinauer Associates, Sunderland, MA
Ganji Moghadam E, Khalighi A (2007) Relationship between vigor of Iranian Prunus mahaleb L. selected dwarf rootstocks and some morphological characters. Sci Hort 111:209–212
Gerlach HK, Stosser R (1998) Sweet cherry cultivar identification using RAPD-derived DNA fine prints. Acta Hort 468:63–69
Hillig KW, Iezzoni AF (1988) Multivariate analysis of a sour cherry germplasm collection. J Am Soc Hortic Sci 1988:928–934
Hjalmarsson I, Ortiz R (2000) In situ and ex situ assessment of morphological and fruit variation in Scandinavian sweet cherry. Sci Hort 85:37–49
Horvath A, Zanetto A (2008) Origin of sour cherry (Prunus cerasus L.) genomes. Acta Hort 795:131–136
Iezzoni AF, Schmidt H, Albertini A (1990) Cherries (Prunus). In: Moore JN, Ballington JR Jr (eds) Genetic Resources of Temperate Fruit and Nut Crops, vol 1. I.S.H.S, Wageningen, pp 111–173
Kadkhodaei S, Elahy M, Khayyam Nekouei M, Imani A, Shahnazari M, Mardi M, Javanmard A, Arbakariya B (2011) A panel of cultivate specific marker based on polymorphisms at microsatellite markers for Iranian cultivated Almonds (Prunus dulcis). AJCS 4:730–736
Khadivi-Khub A, Zamani Z, Bouzari N (2008) Evaluation of genetic diversity in some Iranian and foreign sweet cherry cultivars by using RAPD molecular markers and morphological traits. Hortic Environ Biotechnol 49:188–196
Khadivi-Khub A, Zamani Z, Fatahi MR (2012) Multivariate analysis of Prunus subgen. Cerasus germplasm in Iran using morphological ariables. Genet Resour Crop Evol 59:909–926
Krahl KH, Lansari A, Iezzoni AF (1991) Morphological variation within a sour cherry collection. Euphytica 52:47–55
Kumar LS (1999) DNA markers in plant improvement: an overview. Biotech Advan 17:143–182
Lacis G, Rashal I, Ruisa S, Trajkovski V, Iezzoni AF (2009) Assessment of genetic diversity of Latvian and Swedish sweet cherry (Prunus avium L.) genetic resources collections by using SSR (microsatellite) markers. Sci Hort 121:451–457
Li MM, Cai YL, Qian ZQ, Zhao GF (2009) Genetic diversity and differentiation in Chinese sour cherry Prunus pseudocerasus Lindl., and its implications for conservation. Genet Resour Crop Evol 56:455–464
Linnaeus C (1753) Species Plantarum, 1st edn. Holmiae Marshall RE (1954) Cherries and Cherry Products. In: Economic crops. Interscience, ed., vol 5 New York
Lisek A, Rozpara E (2009) Identification and genetic diversity assessment of cherry cultivars and rootstocks using ISSR-PCR technique. J Fruit Ornam Plant Res 17:95–106
Lisek A, Korbin M, Rozpara E (2006) Using simply generated RAPD markers to distinguish between sweet cherry (Prunus avium L.) cultivars. J Fruit Ornam Plant Res 14:53–59
Miteham EJ, Clayton M, Biasi WV (1998) Comparison of devices for measuring cherry fruit firmness. Hort Sci 33:723–727
Perez R, Navarro F, Sanchez MA, Ortiz JM, Morales R (2010) Analysis of agromorphological descriptors to differentiate between duke cherry (Prunus x gondouinii (Poit. & Turpin) Rehd.) and its progenitors: sweet cherry (Prunus avium L.) and sour cherry (Prunus cerasus L.). Chilean J Agri Res 70:34–49
Perez-Sanchez R, Gomez-Sanchez MA, Morales-Corts R (2008) Agromorphological characterization of traditional Spanish sweet cherry (Prunus avium L.), sour cherry (Prunus cerasus L.) and duke cherry (Prunus x gondouinii Rehd.) cultivars. Span J Agri Res 6:42–55
Powell W, Morgante M, McDevitt R, Vendramin GC, Rafalski JA (1996) Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci USA 92:7759–7763
Rakonjac V, Fotiric Aksic M, Nikolic D, Milatovic D, Colic S (2010) Morphological characterization of ‘Oblacinska’ sour cherry by multivariate analysis. Sci Hort 125:679–684
Rodrigues LC, Morales MR, Fernandes AJB, Ortiz JM (2008) Morphological characterization of sweet and sour cherry cultivars in a germplasm bank at Portugal. Genet Resour Crop Evol 55:593–601
Rohlf FJ (1994) NTSYS-pc, numerical taxonomy and multivariate analysis system 1.80. Exeter Software, New York
Romesburg HC (1990) Cluster Analysis for Researchers. Krieger Publisher, Malabar
Santi F, Lemoine M (1990) Genetic markers for Prunus avium L. 2. Clonal identifications and discrimination from P. cerasus and P. cerasus × P. avium. Annales des Sciences Forestières 47:219–227
SAS (2003) SAS® 9.1 Qualification Tools User’s Guide. SAS Institute Inc., Cary, NC
Schmidt H, Christensen JV, Watkins R, Smith RA (1985) Cherry descriptor list. CEC Secretariat, Brussels. AGPG: IBPGR/85/37, November
Sefc KM, Lopez MS, Lefort F, Botta R (2000) Microsatellites variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor Appl Genet 100:498–505
Semagn K (2002) Genetic relationships among ten encoded types as revealed by a combination of morphological, RAPD and AFLP markers. Hereditas 137:149–156
Shahi-Gharahlar A, Zamani Z, Fatahi R, Bouzari N (2011) Estimation of genetic diversity in some Iranian wild Prunus subgenus Cerasus accessions using inter-simple sequence repeat (ISSR) markers. Biochem Syst and Ecol 39:826–833
Shimada T, Hayama H, Nishimura K, Yamaguchi M, Yoshida M (2001) The genetic diversities of 4 species of subg. Lithocerasus (Prunus, Rosaceae) revealed by RAPD analysis. Euphytica 117:85–90
Sneath PHA, Sokal RR (1973) Numerical Taxonomy. Freeman, San Francisco
Tavaud M, Zanetto A, Santi F, Dirlewanger E (2001) Structuration of genetic diversity in cultivated and wild cherry varieties using molecular markers. Acta Hort 546:263–269
Tavaud M, Zanetto A, David JL, Laigret F, Dirlewanger E (2004) Genetic relationships between diploid and allotetraploid cherry species (Prunus avium, Prunus × gondouinii and Prunus cerasus. Heredity 93:631–638
Turkoglu Z, Bilgener S, Ercisli S, Bakir M, Koc A, Akbulut M, Gercekcioglu R, Gunes M, Esitken A (2010) Simple sequence repeat-based assessment of genetic relationships among Prunus rootstocks. Genetics and Mol Res 9:2156–2165
Upadhyay A, Jayadev K, Manimekalai R, Parthasarathy VA (2004) Genetic relationship and diversity in Indian coconut accessions based on RAPD markers. Sci Hort 99:353–362
Weising K, Nybom H, Wolff K, Gunter K (2005) DNA Fingerprinting in Plants, Principle, Methods, and Applications, 2nd edn. CRC Press, Boca Raton
Williams JGK, Kubelik AE, Livak KJ, Rafaiski JA, Tingey SC (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Res 18:6531–6536
Yarilgac T (2001) Some characteristics of native sour cherry genotypes grown by seed in Van region. Tarim Bilimleri Dergisi 11:13–17
Zamani Z, Shahi-Gharahlar A, Fatahi R, Bouzari N (2012) Genetic relatedness among some wild cherry (Prunus subgenus Cerasus) genotypes native to Iran assayed by morphological traits and random amplified polymorphic DNA analysis. Plant Syst Evol 298:499–509
Zhou L, Kappel F, Wiersma PA, Hampson C, Bakkeren G (2005) Genetic analysis and DNA fingerprinting of sweet cherry cultivars and selections using amplified fragment length polymorphisms (AFLP). Acta Hort 667:37–44
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Carlson.
Rights and permissions
About this article
Cite this article
Khadivi-Khub, A., Jafari, HR. & Zamani, Z. Phenotypic and genotypic variation in Iranian sour and duke cherries. Trees 27, 1455–1466 (2013). https://doi.org/10.1007/s00468-013-0892-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-013-0892-y