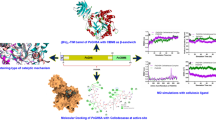
Abstract
Cellobiose 2-epimerase (CE) is ideally suited to synthesize lactulose from lactose, but the poor thermostability and catalytic efficiency restrict enzymatic application. Herein, a non-characterized CE originating from Caldicellulosiruptor morganii (CmCE) was discovered in the NCBI database. Then, a smart mutation library was constructed based on FoldX ΔΔG calculation and modeling structure analysis, from which a positive mutant D226G located within the α8/α9 loop exhibited longer half-lives at 65–75 °C as well as lower Km and higher kcat/Km values compared with CmCE. Molecular modeling demonstrated that the improvement of D226G was largely attributed to the rigidification of the flexible loop, the compactness of the catalysis pocket and the increment of substrate-binding capability. Finally, the yield of synthesizing lactulose catalyzed by D226G reached 45.5%, higher than the 35.9% achieved with CmCE. The disclosed effect of the flexible loop on enzymatic stability and catalysis provides insight to redesign efficient CEs to biosynthesize lactulose.
Graphical abstract








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Guerrero C, Valdivia F, Ubilla C, Ramírez N, Gómez M, Aburto C, Vera C, Illanes A (2019) Continuous enzymatic synthesis of lactulose in packed-bed reactor with immobilized Aspergillus oryzae β-galactosidase. Bioresour Technol 278:296–302. https://doi.org/10.1016/j.biortech.2018.12.018
Sitanggang AB, Drews A, Kraume M (2016) Development of a continuous membrane reactor process for enzyme-catalyzed lactulose synthesis. Biochem Eng J 109:65–80. https://doi.org/10.1016/j.bej.2016.01.006
Nooshkam M, Babazadeh A, Jooyandeh H (2018) Lactulose: properties, techno-functional food applications, and food grade delivery system. Trends Food Sci Technol 80:23–34. https://doi.org/10.1016/j.tifs.2018.07.028
Wang M, Wang L, Lyu X, Hua X, Goddard JM, Yang R (2022) Lactulose production from lactose isomerization by chemo-catalysts and enzymes: current status and future perspectives. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2022.108021
Shen Q, Zhang Y, Yang R, Pan S, Dong J, Fan Y, Han L (2016) Enhancement of isomerization activity and lactulose production of cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus. Food Chem 207:60–67. https://doi.org/10.1016/j.foodchem.2016.02.067
Kuschel B, Claaßen W, Mu W, Jiang B, Stressler T, Fischer L (2016) Reaction investigation of lactulose-producing cellobiose 2-epimerases under operational relevant conditions. J Mol Catal B: Enzym 133:80–87. https://doi.org/10.1016/j.molcatb.2016.11.022
Chen Q, Levin R, Zhang W, Zhang T, Jiang B, Stressler T, Fischer L, Mu W (2017) Characterisation of a novel cellobiose 2-epimerase from thermophilic Caldicellulosiruptor obsidiansis for lactulose production. J Sci Food Agric 97:3095–3105. https://doi.org/10.1002/jsfa.8148
Kim JE, Kim YS, Kang LW, Oh DK (2012) Characterization of a recombinant cellobiose 2-epimerase from Dictyoglomus turgidum that epimerizes and isomerizes β-1,4- and α-1,4-gluco-oligosaccharides. Biotechnol Lett 34:2061–2068. https://doi.org/10.1007/s10529-012-0999-z
Park CS, Kim JE, Choi JG, Oh DK (2011) Characterization of a recombinant cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus and its application in the production of mannose from glucose. Appl Microbiol Biotechnol 92:1187–1196. https://doi.org/10.1007/s00253-011-3403-3
Xiao Y, Chen Q, Shakhnovich EI, Zhang W, Mu W (2019) Simulation-guided enzyme discovery: a new microbial source of cellobiose 2-epimerase. Int J Biol Macromol 139:1002–1008. https://doi.org/10.1016/j.ijbiomac.2019.08.075
Fujiwara T, Saburi W, Matsui H, Mori H, Yao M (2014) Structural insights into the epimerization of β-1,4-Linked oligosaccharides catalyzed by cellobiose 2-epimerase, the sole enzyme epimerizing non-anomeric hydroxyl groups of unmodified sugars. J Biol Chem 289:3405–3415. https://doi.org/10.1074/jbc.m113.531251
Xu G, Kunzendorf A, Crotti M, Rozeboom HJ, Thunnissen AWH, Poelarends GJ (2022) Gene fusion and directed evolution to break structural symmetry and boost catalysis by an oligomeric C−C bond-forming enzyme. Angew Chem Int Ed 61:e202113970. https://doi.org/10.1002/anie.202113970
Shen Q, Zhang Y, Yang R, Hua X, Zhang W, Zhao W (2015) Thermostability enhancement of cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus by site-directed mutagenesis. J Mol Catal B Enzym 120:158–164. https://doi.org/10.1016/j.molcatb.2015.07.007
Feng Y, Lyu X, Cong Y, Hua X, Sun H, Wang L, Yao C, Yang R (2020) Insight into the significant roles of the Trp372 and flexible loop in directing the catalytic direction and substrate specificity in AGE superfamily enzymes. Biochem Eng J 161:107662. https://doi.org/10.1016/j.bej.2020.107662
Wang X, Du J, Zhao B, Wang H, Rao S, Du G, Zhou J, Chen J, Liu S (2021) Significantly improving the thermostability and catalytic efficiency of Streptomyces mobaraenesis transglutaminase through combined rational design. J Agric Food Chem 69:15268–15278. https://doi.org/10.1021/acs.jafc.1c05256
Daudé D, Topham CM, Remaud-Siméon M, André I (2013) Probing impact of active site residue mutations on stability and activity of Neisseria polysaccharea amylosucrase: probing impact of active site residue mutations. Protein Sci 22:1754–1765. https://doi.org/10.1002/pro.2375
Bi J, Chen S, Zhao X, Nie Y, Xu Y (2020) Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl Microbiol Biotechnol 104:7551–7562. https://doi.org/10.1007/s00253-020-10764-z
Buß O, Muller D, Jager S, Rudat J, Rabe KS (2018) Improvement in the thermostability of a β-amino acid converting ω-transaminase by using FoldX. ChemBioChem 19:379–387. https://doi.org/10.1002/cbic.201800465
Wang R, Wang S, Xu Y, Yu X (2020) Enhancing the thermostability of Rhizopus chinensis lipase by rational design and MD simulations. Int J Biol Macromol 160:1189–1200. https://doi.org/10.1016/j.ijbiomac.2020.05.243
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Žídek A, Green T, Tunyasuvunakool K, Petersen S, Jumper J, Clancy E, Green R, Vora A, Lutfi M, Figurnov M, Cowie A, Hobbs N, Kohli P, Kleywegt G, Birney E, Hassabis D, Velankar S (2022) AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444. https://doi.org/10.1093/nar/gkab1061
Wang L, Gu J, Feng Y, Wang M, Tong Y, Liu Y, Lyu X, Yang R (2021) Enhancement of the isomerization activity and thermostability of cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus by exchange of a flexible loop. J Agric Food Chem 69:1907–1915. https://doi.org/10.1021/acs.jafc.0c07073
Jia DX, Zhou L, Zheng YG (2017) Properties of a novel thermostable glucose isomerase mined from Thermus oshimai and its application to preparation of high fructose corn syrup. Enzyme Microb Technol 99:1–8. https://doi.org/10.1016/j.enzmictec.2017.01.001
Xu P, Ni ZF, Zong MH, Ou XY, Yang JG, Lou WY (2020) Improving the thermostability and activity of Paenibacillus pasadenensis chitinase through semi-rational design. Int J Biol Macromol 150:9–15. https://doi.org/10.1016/j.ijbiomac.2020.02.033
Chen A, Li Y, Nie J, McNeil B, Jeffrey L, Yang Y, Bai Z (2015) Protein engineering of Bacillus acidopullulyticus pullulanase for enhanced thermostability using in silico data driven rational design methods. Enzyme Microb Technol 78:74–83. https://doi.org/10.1016/j.enzmictec.2015.06.013
Chen Q, He W, Yan X, Zhang T, Jiang B, Stressler T, Fischer L, Mu W (2018) Construction of an enzymatic route using a food-grade recombinant Bacillus subtilis for the production and purification of epilactose from lactose. J Dairy Sci 101:1872–1882. https://doi.org/10.3168/jds.2017-12936
Feng Y, Hua X, Shen Q, Matthews M, Zhang Y, Fisher AJ, Lyu X, Yang R (2020) Insight into the potential factors influencing the catalytic direction in cellobiose 2-epimerase by crystallization and mutagenesis. Acta Crystallogr Sect D Struct Biol 76:1104–1113. https://doi.org/10.1107/s205979832001222x
Xu Z, Cen YK, Zou SP, Xue YP, Zheng YG (2020) Recent advances in the improvement of enzyme thermostability by structure modification. Crit Rev Biotechnol 40:83–98. https://doi.org/10.1080/07388551.2019.1682963
Caldararu O, Blundell TL, Kepp KP (2021) A base measure of precision for protein stability predictors: structural sensitivity. BMC Bioinf 22:88. https://doi.org/10.1186/s12859-021-04030-w
Thiltgen G, Goldstein RA (2012) Assessing predictors of changes in protein stability upon mutation using self-consistency. PLoS ONE 7:e46084. https://doi.org/10.1371/journal.pone.0046084
Jameson JK, Mathiesen G, Pope PB, Westereng B, La Rosa SL (2021) Biochemical characterization of two cellobiose 2-epimerases and application for efficient production of lactulose and epilactose. Current Res Biotechnol 3:57–64. https://doi.org/10.1016/j.crbiot.2021.02.003
Rentschler E, Schuh K, Krewinkel M, Baur C, Claaßen W, Meyer S, Kuschel B, Stressler T, Fischer L (2015) Enzymatic production of lactulose and epilactose in milk. J Dairy Sci 98:6767–6775. https://doi.org/10.3168/jds.2015-9900
Kim YS, Oh DK (2012) Lactulose production from lactose as a single substrate by a thermostable cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus. Bioresour Technol 104:668–672. https://doi.org/10.1016/j.biortech.2011.11.016
Nguyen C, Young JT, Slade GG, Oliveira RJ, McCully ME (2019) A dynamic hydrophobic core and surface salt bridges thermostabilize a designed three-helix bundle. Biophys J 116:621–632. https://doi.org/10.1016/j.bpj.2019.01.012
Lee JM, Moon SY, Kim YR, Kim KW, Lee BJ, Kong IS (2017) Improvement of thermostability and halostability of β-1,3–1,4-glucanase by substituting hydrophobic residue for Lys 48. Int J Biol Macromol 94:594–602. https://doi.org/10.1016/j.ijbiomac.2016.10.043
Zhang H, Sang J, Zhang Y, Sun T, Liu H, Yue R, Zhang J, Wang H, Dai Y, Lu F, Liu F (2019) Rational design of a Yarrowia lipolytica derived lipase for improved thermostability. Int J Biol Macromol 137:1190–1198. https://doi.org/10.1016/j.ijbiomac.2019.07.070
Jia DX, Wang F, Zhao R, Gu BD, Peng C, Jin LQ, Liu ZQ, Zheng YG (2022) Engineering novel (R)-selective transaminase for efficient symmetric synthesis of d-alanine. Appl Environ Microbiol 88:e00062-e122. https://doi.org/10.1128/aem.00062-22
Qian H, Zhang C, Lu Z, Xia B, Bie X, Zhao H, Lu F, Yang GY (2018) Consensus design for improved thermostability of lipoxygenase from Anabaena sp. PCC 7120. BMC Biotechnol 18:57. https://doi.org/10.1186/s12896-018-0468-4
Maiello F, Gallo G, Coelho C, Sucharski F, Hardy L, Würtele M (2020) Crystal structure of Thermus thermophilus methylenetetrahydrofolate dehydrogenase and determinants of thermostability. PLoS ONE 15:e0232959. https://doi.org/10.1371/journal.pone.0232959
Reetz MT, Soni P, Acevedo JP, Sanchis J (2009) Creation of an amino acid network of structurally coupled residues in the directed evolution of a thermostable enzyme. Angew Chem Int Ed 121:8418–8422. https://doi.org/10.1002/anie.200904209
Li T, Yang S, Wang X, Cai H, Wang Y, Li C, Li E (2022) Improving thermostability of GH11 xylanase xynASP by the design of loop region. Crystals 12:1228. https://doi.org/10.3390/cryst12091228
Zhou Y, Jiao L, Shen J, Chi H, Lu Z, Liu H, Lu F, Zhu P (2022) Enhancing the catalytic activity of type II L-asparaginase from Bacillus licheniformis through semi-rational design. Int J Mol Sci 23:9663. https://doi.org/10.3390/ijms23179663
Gong XM, Qin Z, Li FL, Zeng BB, Zheng GW, Xu JH (2019) Development of an engineered ketoreductase with simultaneously improved thermostability and activity for making a eulky atorvastatin precursor. ACS Catal 9:147–153. https://doi.org/10.1021/acscatal.8b03382
Kim JH, Choi GS, Kim SB, Kim WH, Lee JY, Ryu YW, Kim GJ (2004) Enhanced thermostability and tolerance of high substrate concentration of an esterase by directed evolution. J Mol Catal B Enzym 27:169–175. https://doi.org/10.1016/j.molcatb.2003.11.010
Park AR, Kim JS, Jang SW, Park YG, Koo BS, Lee HC (2017) Rational modification of substrate-binding site by structure-based engineering of a cellobiose 2-epimerase in Caldicellulosiruptor saccharolyticus. Microb Cell Factories 16:224. https://doi.org/10.1186/s12934-017-0841-3
Acknowledgements
This work was supported by the Program of the National Key Research and Development Project (2020YFA0908400), the Pioneer R&D Program of Zhejiang (2023C01113), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang, P. R. China (2018R01014), the Fundamental Research Funds for the Provincial Universities of Zhejiang (RF-C2019005), and the Key Research and Development Program of Zhejiang Province (2021C03093).
Author information
Authors and Affiliations
Contributions
D-XJ: conceptualization, data curation, writing—original draft, and writing—review & editing. HY: methodology, software, validation, investigation, and writing—original draft. FW: methodology, software, validation, investigation, and writing—original draft. L-QJ: visualization, and writing—original draft. Z-QL: visualization and supervision. Y-GZ: supervision and project administration.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jia, DX., Yu, H., Wang, F. et al. Computer-aided design of novel cellobiose 2-epimerase for efficient synthesis of lactulose using lactose. Bioprocess Biosyst Eng 46, 1279–1291 (2023). https://doi.org/10.1007/s00449-023-02896-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-023-02896-z