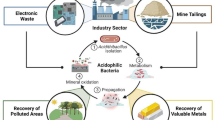
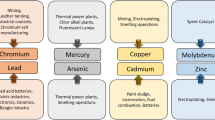
Abstract
Acidophiles are a group of microorganisms that thrive in acidic environments where pH level is far below the neutral value 7.0. They belong to a larger family called extremophiles, which is a group that thrives in various extreme environmental conditions which are normally inhospitable to other organisms. Several human activities such as mining, construction and other industrial processes release highly acidic effluents and wastes into the environment. Those acidic wastes and wastewaters contain different types of pollutants such as heavy metals, radioactive, and organic, whose have adverse effects on human being as well as on other living organisms. To protect the whole ecosystem, those pollutants containing effluents or wastes must be clean properly before releasing into environment. Physicochemical cleanup processes under extremely acidic conditions are not always successful due to high cost and release of toxic byproducts. While in case of biological methods, except acidophiles, no other microorganisms cannot survive in highly acidic conditions. Therefore, acidophiles can be a good choice for remediation of different types of contaminants present in acidic conditions. In this review article, various roles of acidophilic microorganisms responsible for removing heavy metals and radioactive pollutants from acidic environments were discussed. Bioremediation of various acidic organic pollutants by using acidophiles was also studied. Overall, this review could be helpful to extend our knowledge as well as to do further relevant novel studies in the field of acidic pollutants remediation by applying acidophilic microorganisms.




Similar content being viewed by others
Data availability statement
No data was used for the research described in the article.
References
Ali N, Dashti N, Khanafer M et al (2020) Bioremediation of soils saturated with spilled crude oil. Sci Rep 10:1–9. https://doi.org/10.1038/s41598-019-57224-x
Rambabu K, Banat F, Pham QM et al (2020) Biological remediation of acid mine drainage: Review of past trends and current outlook. Environ Sci Ecotechnol 2:100024. https://doi.org/10.1016/j.ese.2020.100024
Cleary A, Lloyd JR, Newsome L et al (2019) Bioremediation of strontium and technetium contaminated groundwater using glycerol phosphate. Chem Geol 509:213–222. https://doi.org/10.1016/j.chemgeo.2019.02.004
Adnan LA, Sathishkumar P, Rahim A, Yusoff M (2017) Rapid bioremediation of Alizarin Red S and Quinizarine Green SS dyes using Trichoderma lixii F21 mediated by biosorption and enzymatic processes. Bioprocess Biosyst Eng 40:85–97. https://doi.org/10.1007/s00449-016-1677-7
Andreas HT, Sathishkumar P et al (2021) Microplastic contamination in the Skipjack Tuna (Euthynnus affinis) collected from Southern Coast of Java, Indonesia. Chemosphere 276:130185. https://doi.org/10.1016/J.CHEMOSPHERE.2021.130185
Sathishkumar P, Mohan K, Ganesan AR et al (2021) Persistence, toxicological effect and ecological issues of endosulfan—a review. J Hazard Mater 416:125779. https://doi.org/10.1016/J.JHAZMAT.2021.125779
Hui Li AS, Sathishkumar P, Selahuddeen ML et al (2022) Adverse environmental effects of disposable face masks due to the excess usage. Environ Pollut 308:119674. https://doi.org/10.1016/J.ENVPOL.2022.119674
Fashola M, Ngole-Jeme V, Babalola O (2015) Diversity of acidophilic bacteria and archaea and their roles in bioremediation of acid mine drainage. Br Microbiol Res J 8:443–456. https://doi.org/10.9734/bmrj/2015/14365
Taylor AJ, Pape S, Murphy N (2005) A summary of passive and active treatment technologies for acid and metalliferous drainage (AMD). Australian Centre for Minerals Extension and Research (ACMER), Fifth Australian Workshop on Acid Drainage, 29-31 August 2005, Fremantle, Australia
Jeong SW, Choi YJ (2020) Extremophilic microorganisms for the treatment of toxic pollutants in the environment. Molecules 25:13–15. https://doi.org/10.3390/molecules25214916
Zhang C, Wu D, Ren H (2020) Bioremediation of oil contaminated soil using agricultural wastes via microbial consortium. Sci Rep 10:1–8. https://doi.org/10.1038/s41598-020-66169-5
Dixit R, Wasiullah MD et al (2015) Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustain 7:2189–2212. https://doi.org/10.3390/su7022189
Diep P, Mahadevan R, Yakunin AF (2018) Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2018.00157
Igiri BE, Okoduwa SIR, Idoko GO et al (2018) Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J Toxicol. https://doi.org/10.1155/2018/2568038
Kiadehi MSH, Amoozegar MA, Asad S, Siroosi M (2018) Exploring the potential of halophilic archaea for the decolorization of azo dyes. Water Sci Technol 77:1602–1611. https://doi.org/10.2166/wst.2018.040
Tkavc R, Matrosova VY, Grichenko OE et al (2018) Prospects for fungal bioremediation of acidic radioactive waste sites: characterization and genome sequence of Rhodotorula taiwanensis MD1149. Front Microbiol 8:1–21. https://doi.org/10.3389/fmicb.2017.02528
Angelov A, Liebl W (2006) Insights into extreme thermoacidophily based on genome analysis of Picrophilus torridus and other thermoacidophilic archaea. J Biotechnol 126:3–10. https://doi.org/10.1016/j.jbiotec.2006.02.017
Siliakus MF, van der Oost J, Kengen SWM (2017) Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 21:651–670. https://doi.org/10.1007/s00792-017-0939-x
Johnson DB, Quatrini R (2020) Acidophile microbiology in space and time. Curr Issues Mol Biol 39:63–76. https://doi.org/10.21775/cimb.039.063
Krulwich TASGPE (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. https://doi.org/10.1038/nrmicro2549
Navarro CA, von Bernath D, Jerez CA (2013) Heavy metal resistance strategies of acidophilic bacteria and their acquisition: importance for biomining and bioremediation. Biol Res 46:363–371
Chen X (2021) Thriving at low pH: adaptation mechanisms of acidophiles. Acidophiles Fundam Appl (Working Title). https://doi.org/10.5772/INTECHOPEN.96620
Mirete S, Morgante V, González-Pastor JE (2017) Acidophiles: diversity and mechanisms of adaptation to acidic environments. In: Adaption of microbial life to environmental extremes: novel research results and application, 2nd edn. Springer, London, pp 227–251
Sharma A, Parashar D, Satyanarayana T (2016) Acidophilic microbes: biology and applications, pp 215–241. https://doi.org/10.1007/978-3-319-13521-2_7
Pöhler I, Wenderoth D, Wendt-Potthoff K, Höfle M (2002) Bacterioplankton community structure and dynamics in enclosures during bioremediation experiments in an acid mining lake. Water Air Soil Pollut Focus 2:111–121. https://doi.org/10.1023/A:1019951612385
Johnson DB, Hallberg KB (2003) The microbiology of acidic mine waters. Res Microbiol 154:466–473. https://doi.org/10.1016/S0923-2508(03)00114-1
Johnson DB (2012) Geomicrobiology of extremely acidic subsurface environments. FEMS Microbiol Ecol 81:2–12. https://doi.org/10.1111/j.1574-6941.2011.01293.x
Ayangbenro AS, Olanrewaju OS, Babalola OO (2018) Sulfate-reducing bacteria as an effective tool for sustainable acid mine bioremediation. Front Microbiol 9:1–10. https://doi.org/10.3389/fmicb.2018.01986
Hassan N, Rafiq M, Rehman M et al (2019) Fungi in acidic fire: a potential source of industrially important enzymes. Fungal Biol Rev 33:58–71
Panda S, Mishra S, Akcil A (2016) Bioremediation of acidic mine effluents and the role of sulfidogenic biosystems: a mini-review. Euro-Mediterranean J Environ Integr 1:1–9. https://doi.org/10.1007/s41207-016-0008-3
Hallberg KB (2010) New perspectives in acid mine drainage microbiology. Hydrometallurgy 104:448–453. https://doi.org/10.1016/j.hydromet.2009.12.013
Hedrich S, Schippers A (2020) Distribution of acidophilic microorganisms in natural and man-made acidic environments. Curr Issues Mol Biol 40:25–48. https://doi.org/10.21775/CIMB.040.025
Schippers A, Kock D (2009) Geomicrobiology of sulfidic mine dumps: a short review. Adv Mater Res 71–73:37–41. https://doi.org/10.4028/www.scientific.net/AMR.71-73.37
Vaiopoulou E, Provijn T, Prévoteau A et al (2016) Electrochemical sulfide removal and caustic recovery from spent caustic streams. Water Res 92:38–43. https://doi.org/10.1016/j.watres.2016.01.039
Sivasubramanian V, Subramanian VV, Raghavan BG, Ranjithkumar R (2009) Large scale phycoremediation of acidic effluent from an alginate industry. ScienceAsia 35:220–226. https://doi.org/10.2306/scienceasial513-1874.2009.35.220
Phyo AK, Jia Y, Tan Q et al (2020) Competitive growth of sulfate-reducing bacteria with bioleaching acidophiles for bioremediation of heap bioleaching residue. Int J Environ Res Public Health 17:1–14. https://doi.org/10.3390/ijerph17082715
Ayangbenro AS, Babalola OO (2017) A new strategy for heavy metal polluted environments : a review of microbial biosorbents. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph14010094
Sheoran AS, Sheoran V (2006) Heavy metal removal mechanism of acid mine drainage in wetlands: a critical review. Miner Eng 19:105–116. https://doi.org/10.1016/j.mineng.2005.08.006
Volesky B (2001) Detoxification of metal-bearing effluents: biosorption for the next century. Hydrometallurgy 59:203–216. https://doi.org/10.1016/S0304-386X(00)00160-2
Gadd GM (2004) Microbial influence on metal mobility and application for bioremediation. Geoderma 122:109–119. https://doi.org/10.1016/j.geoderma.2004.01.002
McCullough CD, Lund MA (2011) Bioremediation of Acidic and Metalliferous Drainage (AMD) through organic carbon amendment by municipal sewage and green waste. J Environ Manag 92:2419–2426. https://doi.org/10.1016/j.jenvman.2011.04.011
Sánchez-Andrea I, Sanz JL, Bijmans MFM, Stams AJM (2014) Sulfate reduction at low pH to remediate acid mine drainage. J Hazard Mater 269:98–109. https://doi.org/10.1016/j.jhazmat.2013.12.032
Johnson DB, Hallberg KB (2005) Acid mine drainage remediation options: a review. Sci Total Environ 338:3–14. https://doi.org/10.1016/j.scitotenv.2004.09.002
Natarajan KA (2008) Microbial aspects of acid mine drainage and its bioremediation. Trans Nonferrous Met Soc China (Engl Ed) 18:1352–1360. https://doi.org/10.1016/S1003-6326(09)60008-X
Bridge TAM, Barrie Johnson D (2000) Reductive dissolution of ferric iron minerals by Acidiphilium sjh. Geomicrobiol J 17:193–206. https://doi.org/10.1080/01490450050121161
Malik L, Hedrich S (2022) Ferric iron reduction in extreme acidophiles. Front Microbiol 12:1–17. https://doi.org/10.3389/fmicb.2021.818414
Johnson DB, Kanao T, Hedrich S (2012) Redox transformations of iron at extremely low pH: fundamental and applied aspects. Front Microbiol 3:1–13. https://doi.org/10.3389/fmicb.2012.00096
Luptakova A, Kusnierova M (2005) Bioremediation of acid mine drainage contaminated by SRB. Hydrometallurgy 77:97–102. https://doi.org/10.1016/j.hydromet.2004.10.019
Castro HF, Williams NH, Ogram A (2006) Phylogeny of sulfate-reducing bacteria1. FEMS Microbiol Ecol 31:1–9. https://doi.org/10.1111/j.1574-6941.2000.tb00665.x
Rabus R, Hansen T, Widdel R (2013) Dissimilatory sulfate- and sulfur-reducing prokaryotes. In: The prokaryotes—prokaryotic physiology and biochemistry, pp 309–404
Alexandrino M, Macías F, Costa R et al (2011) A bacterial consortium isolated from an Icelandic fumarole displays exceptionally high levels of sulfate reduction and metals resistance. J Hazard Mater 187:362–370. https://doi.org/10.1016/j.jhazmat.2011.01.035
Sánchez-Andrea I, Stams AJM, Hedrich S et al (2015) Desulfosporosinus acididurans sp. nov.: an acidophilic sulfate-reducing bacterium isolated from acidic sediments. Extremophiles 19:39–47. https://doi.org/10.1007/s00792-014-0701-6
Hauri JF, Schaider LA (2009) Remediation of acid mine drainage with sulfate reducing bacteria. J Chem Educ 86:216–218. https://doi.org/10.1021/ed086p216
Neculita C-M, Zagury GJ, Bussière B (2007) Passive treatment of acid mine drainage in bioreactors using sulfate-reducing bacteria. J Environ Qual 36:1–16. https://doi.org/10.2134/jeq2006.0066
Bai H, Kang Y, Quan H et al (2013) Treatment of acid mine drainage by sulfate reducing bacteria with iron in bench scale runs. Bioresour Technol 128:818–822. https://doi.org/10.1016/j.biortech.2012.10.070
Hedrich S, Johnson DB (2014) Remediation and selective recovery of metals from acidic mine waters using novel modular bioreactors. Environ Sci Technol. https://doi.org/10.1021/es5030367
Chen LX, Huang LN, Méndez-García C et al (2016) Microbial communities, processes and functions in acid mine drainage ecosystems. Curr Opin Biotechnol 38:150–158. https://doi.org/10.1016/j.copbio.2016.01.013
Sun R, Zhang L, Wang X et al (2020) Elemental sulfur-driven sulfidogenic process under highly acidic conditions for sulfate-rich acid mine drainage treatment: performance and microbial community analysis. Water Res. https://doi.org/10.1016/j.watres.2020.116230
Qiu Y, Guo J, Zhang L et al (2017) A high-rate sulfidogenic process based on elemental sulfur reduction: cost-effectiveness evaluation and microbial community analysis a high-rate sulfidogenic process based on elemental sulfur reduction: cost-effectiveness evaluation and microbial community analysis. Biochem Eng J. https://doi.org/10.1016/j.bej.2017.09.001
Sun R, Zhang L, Zhang Z et al (2018) Realizing high-rate sulfur reduction under sulfate-rich conditions in a biological sul fi de production system to treat metal-laden wastewater de fi cient in organic matter. Water Res 131:239–245. https://doi.org/10.1016/j.watres.2017.12.039
Guo J, Wang J, Qiu Y et al (2019) Chemosphere Realizing a high-rate sul fi dogenic reactor driven by sulfur-reducing bacteria with organic substrate dosage minimization and cost-effectiveness maximization. Chemosphere 236:124381. https://doi.org/10.1016/j.chemosphere.2019.124381
Florentino AP, Weijma J, Stams AJM, Sánchez-Andrea I (2015) Sulfur reduction in acid rock drainage environments. Environ Sci Technol 49:11746–11755. https://doi.org/10.1021/acs.est.5b03346
Sánchez-Andrea I, Rodríguez N, Amils R, Sanz JL (2011) Microbial diversity in anaerobic sediments at Río Tinto, a naturally acidic environment with a high heavy metal content. Appl Environ Microbiol 77:6085–6093. https://doi.org/10.1128/AEM.00654-11
Kaksonen AH, Plumb JJ, Franzmann PD, Puhakka JA (2004) Simple organic electron donors support diverse sulfate-reducing communities in fluidized-bed reactors treating acidic metal- and sulfate-containing wastewater. FEMS Microbiol Ecol 47:279–289. https://doi.org/10.1016/S0168-6496(03)00284-8
Johnson DB (1995) Acidophilic microbial communities: candidates for bioremediation of acidic mine effluents. Int Biodeterior Biodegrad 35:41–58. https://doi.org/10.1016/0964-8305(95)00065-D
Emerson D, Moyer C (1997) Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol 63:4784–4792. https://doi.org/10.1128/aem.63.12.4784-4792.1997
Bwapwa JK, Jaiyeola AT, Chetty R (2017) Bioremediation of acid mine drainage using algae strains: a review. South Afr J Chem Eng 24:62–70. https://doi.org/10.1016/j.sajce.2017.06.005
Dean AP, Hartley A, McIntosh OA et al (2019) Metabolic adaptation of a Chlamydomonas acidophila strain isolated from acid mine drainage ponds with low eukaryotic diversity. Sci Total Environ 647:75–87. https://doi.org/10.1016/j.scitotenv.2018.07.445
Chen S, Lin J (2001) Effect of substrate concentration on bioleaching of metal-contaminated sediment. J Hazardous Mater 82:77–89
Beolchini F, Dell’Anno A, De Propris L et al (2009) Auto- and heterotrophic acidophilic bacteria enhance the bioremediation efficiency of sediments contaminated by heavy metals. Chemosphere 74:1321–1326. https://doi.org/10.1016/j.chemosphere.2008.11.057
Küsel K, Roth U, Drake HL (2002) Microbial reduction of Fe(III) in the presence of oxygen under low pH conditions. Environ Microbial 4:414–421
Cabrera G, Pérez R, Gómez JM et al (2006) Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp. strains. J Hazard Mater 135:40–46. https://doi.org/10.1016/j.jhazmat.2005.11.058
Marques CR (2018) Extremophilic microfactories: applications in metal and radionuclide bioremediation. Front Microbiol 9:1–10. https://doi.org/10.3389/fmicb.2018.01191
Li J, Zhang Y (2012) Remediation technology for the uranium contaminated environment: a review. Procedia Environ Sci 13:1609–1615. https://doi.org/10.1016/j.proenv.2012.01.153
Voica DM, Bartha L, Banciu HL, Oren A (2016) Heavy metal resistance in halophilic Bacteria and Archaea. FEMS Microbiol Lett 363:1–9. https://doi.org/10.1093/femsle/fnw146
Sand W, Gehrke T (2006) Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res Microbiol 157:49–56. https://doi.org/10.1016/j.resmic.2005.07.012
Llorens I, Untereiner G, Jaillard D et al (2012) Uranium interaction with two multi-resistant environmental bacteria: Cupriavidus metallidurans CH34 and Rhodopseudomonas palustris. PLoS ONE 7:1–9. https://doi.org/10.1371/journal.pone.0051783
Orell A, Navarro CA, Rivero M et al (2012) Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 16:573–583. https://doi.org/10.1007/s00792-012-0457-9
Brim H, Mcfarlan SC, Fredrickson JK et al (2000) Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat Biotechnol 308:85–90
Daly MJ (2000) Engineering radiation-resistant bacteria for environmental. Curr Opin Biotechnol 11:280–285
Carvalho FP (2017) Mining industry and sustainable development: time for change. Food Energy Secur 6:61–77. https://doi.org/10.1002/fes3.109
Marques CR, Caetano AL, Haller A et al (2014) Toxicity screening of soils from different mine areas—a contribution to track the sensitivity and variability of Arthrobacter globiformis assay. J Hazardous Mater 274:331–341. https://doi.org/10.1016/j.jhazmat.2014.03.066
Carvalho RPSBFP (2014) Uranium mining in Portugal: a review of the environmental legacies of the largest mines and environmental and human health impacts. Environ Geochem Health 36:285–301. https://doi.org/10.1007/s10653-013-9563-6
Venkateswarlu K, Nirola R, Kuppusamy S (2016) Abandoned metalliferous mines: ecological impacts and potential approaches for reclamation. Rev Environ Sci Bio/Technol 15:327–354. https://doi.org/10.1007/s11157-016-9398-6
Gupta P, Diwan B (2017) Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies. Biotechnol Reports 13:58–71. https://doi.org/10.1016/j.btre.2016.12.006
Shukla A, Parmar P, Saraf M (2017) Radiation, radionuclides and bacteria: an in-perspective review. J Environ Radioact 180:27–35. https://doi.org/10.1016/j.jenvrad.2017.09.013
Anderson RT, Vrionis HA, Ortiz-Bernad I et al (2003) Stimulating the in situ activity of. Society 69:5884–5891. https://doi.org/10.1128/AEM.69.10.5884
Nancucheo I, Johnson DB (2011) Significance of microbial communities and Interactions in Safeguarding reactive mine tailings by ecological engineering. Appl Environ Microbiol 77:8201–8208. https://doi.org/10.1128/AEM.06155-11
Lloyd JR, Lovley DR (2001) Microbial detoxification of metals and radionuclides. Curr Opin Biotechnol 12:248–253. https://doi.org/10.1016/S0958-1669(00)00207-X
Ramos JL, Marqués S, van Dillewijn P et al (2011) Laboratory research aimed at closing the gaps in microbial bioremediation. Trends Biotechnol 29:641–647. https://doi.org/10.1016/j.tibtech.2011.06.007
Elleuche S, Schröder C, Sahm K, Antranikian G (2014) Extremozymes-biocatalysts with unique properties from extremophilic microorganisms. Curr Opin Biotechnol 29:116–123. https://doi.org/10.1016/j.copbio.2014.04.003
Raddadi N, Cherif A, Daffonchio D et al (2015) Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl Microbiol Biotechnol 99:7907–7913. https://doi.org/10.1007/s00253-015-6874-9
Dekker L, Arsène-Ploetze F, Santini JM (2016) Comparative proteomics of Acidithiobacillus ferrooxidans grown in the presence and absence of uranium. Res Microbiol 167:234–239. https://doi.org/10.1016/j.resmic.2016.01.007
Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA (2009) Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55:1–79. https://doi.org/10.1016/S0065-2911(09)05501-5
Zhang X, Liu X, Liang Y et al (2016) Metabolic diversity and adaptive mechanisms of iron- and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Environ Microbiol Rep 8:738–751. https://doi.org/10.1111/1758-2229.12435
Takahashi S, Okada H, Abe K, Kera Y (2014) Genetic transformation of the yeast Rhodotorula gracilis ATCC 26217 by electroporation. Appl Biochem Microbiol 50:624–628. https://doi.org/10.1134/S0003683814110040
Su X, Zhou M, Hu P et al (2019) Whole-genome sequencing of an acidophilic Rhodotorula sp. ZM1 and its phenol-degrading capability under acidic conditions. Chemosphere 232:76–86. https://doi.org/10.1016/j.chemosphere.2019.05.195
Masaki Y, Hirajima T, Sasaki K, Okibe N (2015) Bioreduction and immobilization of hexavalent chromium by the extremely acidophilic Fe(III)-reducing bacterium Acidocella aromatica strain PFBC. Extremophiles 19:495–503. https://doi.org/10.1007/s00792-015-0733-6
Amoozegar MA, Ghasemi A, Razavi MR, Naddaf S (2007) Evaluation of hexavalent chromium reduction by chromate-resistant moderately halophile, Nesterenkonia sp. strain MF2. Process Biochem 42:1475–1479. https://doi.org/10.1016/j.procbio.2007.07.001
Amoozegar MA, Ashengroph M, Malekzadeh F et al (2008) Isolation and initial characterization of the tellurite reducing moderately halophilic bacterium, Salinicoccus sp. strain QW6. Microbiol Res 163:456–465. https://doi.org/10.1016/j.micres.2006.07.010
Amoozegar MA, Khoshnoodi M, Didari M et al (2012) Tellurite removal by a tellurium-tolerant halophilic bacterial strain, Thermoactinomyces sp. QS-2006. Ann Microbiol 62:1031–1037. https://doi.org/10.1007/s13213-011-0343-1
Das S, Dash HR, Chakraborty J (2016) Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl Microbiol Biotechnol 100:2967–2984. https://doi.org/10.1007/s00253-016-7364-4
Appukuttan D, Seetharam C, Padma N et al (2011) PhoN-expressing, lyophilized, recombinant Deinococcus radiodurans cells for uranium bioprecipitation. J Biotechnol 154:285–290. https://doi.org/10.1016/j.jbiotec.2011.05.002
Navarro CA, Orellana LH, Mauriaca C, Jerez CA (2009) Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper. Appl Environ Microbiol 75:6102–6109. https://doi.org/10.1128/AEM.00308-09
Cárdenas JP, Valdés J, Quatrini R et al (2010) Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl Microbiol Biotechnol 88:605–620. https://doi.org/10.1007/s00253-010-2795-9
Rojas LA, Yáñez C, González M et al (2011) Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS ONE. https://doi.org/10.1371/journal.pone.0017555
Foo KY, Hameed BH (2009) An overview of landfill leachate treatment via activated carbon adsorption process. J Hazard Mater 171:54–60. https://doi.org/10.1016/j.jhazmat.2009.06.038
Zhou Z, Liu Y, Zanaroli G et al (2019) Enhancing bioremediation potential of pseudomonas putida by developing its acid stress tolerance with glutamate decarboxylase dependent system and global regulator of extreme radiation resistance. Front Microbiol 10:1–8. https://doi.org/10.3389/fmicb.2019.02033
Margesin R, Schinner F (2001) Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl Microbiol Biotechnol 56:650–663. https://doi.org/10.1007/s002530100701
Difference Between Biodegradation and Bioremediation | Environmental Microbiology | The Biology Notes. https://thebiologynotes.com/biodegradation-and-bioremediation/. Accessed 25 July 2021
Parthipan P, Preetham E, Machuca LL, et al (2017) Biosurfactant and degradative enzymes mediated crude oil degradation by bacterium Bacillus subtilis A1. Front Microbiol 8:1–14. https://doi.org/10.3389/fmicb.2017.00193
Gemmell RT, Knowles CJ (2000) Utilisation of aliphatic compounds by acidophilic heterotrophic bacteria. The potential for bioremediation of acidic wastewaters contaminated with toxic organic compounds and heavy metals. FEMS Microbiol Lett 192:185–190. https://doi.org/10.1016/S0378-1097(00)00428-6
Alexander B, Leach S, Ingledew WJ (1987) The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. J Gen Microbiol 133:1171–1179. https://doi.org/10.1099/00221287-133-5-1171
Kishimoto N, Kosako Y, Wakao N et al (1995) Transfer of Acidiphilium facilis and Acidiphilium aminolytica to the Genus Acidocella gen. nov., and Emendation of the Genus Acidiphilium. Syst Appl Microbiol 18:85–91. https://doi.org/10.1016/S0723-2020(11)80453-4
Elumalai P, Parthipan P, Huang M et al (2021) Enhanced biodegradation of hydrophobic organic pollutants by the bacterial consortium: impact of enzymes and biosurfactants. Environ Pollut 289:117956. https://doi.org/10.1016/J.ENVPOL.2021.117956
Röling WFM, Ortega-Lucach S, Larter SR, Head IM (2006) Acidophilic microbial communities associated with a natural, biodegraded hydrocarbon seepage. J Appl Microbiol 101:290–299. https://doi.org/10.1111/j.1365-2672.2006.02926.x
Teske A, Hinrichs KU, Edgcomb V et al (2002) Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68:1994–2007. https://doi.org/10.1128/AEM.68.4.1994-2007.2002
Kniemeyer O, Fischer T, Wilkes H et al (2003) Kniemeyer_2003.pdf. 69:760–768. https://doi.org/10.1128/AEM.69.2.760
Hamamura N, Olson SH, Ward DM, Inskeep WP (2005) Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl Environ Microbiol 71:5943–5950. https://doi.org/10.1128/AEM.71.10.5943-5950.2005
Sood N, Patle S, Lal B (2010) Bioremediation of acidic oily sludge-contaminated soil by the novel yeast strain Candida digboiensis TERI ASN6. Environ Sci Pollut Res 17:603–610. https://doi.org/10.1007/s11356-009-0239-9
Muthukumar B, Parthipan P, AlSalhi MS et al (2022) Characterization of bacterial community in oil-contaminated soil and its biodegradation efficiency of high molecular weight (>C40) hydrocarbon. Chemosphere 289:133168. https://doi.org/10.1016/J.CHEMOSPHERE.2021.133168
Muthukumar B, Al Salhi MS, Narenkumar J et al (2022) Characterization of two novel strains of Pseudomonas aeruginosa on biodegradation of crude oil and its enzyme activities. Environ Pollut 304:119223. https://doi.org/10.1016/J.ENVPOL.2022.119223
Mishra S, Jyot J, Kuhad RC, Lal B (2001) In situ bioremediation potential of an oily sludge-degrading bacterial consortium. Curr Microbiol 43:328–335. https://doi.org/10.1007/s002840010311
Bhattacharya D, Sarma PM, Krishnan S et al (2003) Evaluation of genetic diversity among Pseudomonas citronellolis strains isolated from oily sludge-contaminated sites. Appl Environ Microbiol 69:1435–1441. https://doi.org/10.1128/AEM.69.3.1435-1441.2003
Yuste L, Corbella ME, Turiégano MJ et al (2000) Characterization of bacterial strains able to grow on high molecular mass residues from crude oil processing. FEMS Microbiol Ecol 32:69–75. https://doi.org/10.1016/S0168-6496(00)00015-5
Kloos K, Schloter M, Meyer O (2006) Microbial activity in an acid resin deposit: biodegradation potential and ecotoxicology in an extremely acidic hydrocarbon contamination. Environ Pollut 144:136–144. https://doi.org/10.1016/j.envpol.2005.12.022
Prasad GS, Mayilraj S, Sood N et al (2005) Candida digboiensis sp. nov., a novel anamorphic yeast species from an acidic tar sludge-contaminated oilfield. Int J Syst Evol Microbiol 55:967–972. https://doi.org/10.1099/ijs.0.63313-0
Sood N, Lal B (2009) Isolation of a novel yeast strain Candida digboiensis TERI ASN6 capable of degrading petroleum hydrocarbons in acidic conditions. J Environ Manag 90:1728–1736. https://doi.org/10.1016/j.jenvman.2008.11.026
Parthipan P, Cheng L, Dhandapani P et al (2022) Impact of biosurfactant and iron nanoparticles on biodegradation of polyaromatic hydrocarbons (PAHs). Environ Pollut 306:119384. https://doi.org/10.1016/J.ENVPOL.2022.119384
Al PAK, Huda SQ, Basahi JJGJM (2017) Biodegradation of polycyclic aromatic hydrocarbons by an acidophilic Stenotrophomonas maltophilia strain AJH1 isolated from a mineral mining site in Saudi Arabia. Extremophiles 21:163–174. https://doi.org/10.1007/s00792-016-0892-0
Cerniglia CE (1992) biodegradation of PAHs. Biodegradation 3:351–368
Stapleton RD, Savage DC, Sayler GS, Stacey G (1998) Biodegradation of aromatic hydrocarbons in an extremely acidic environment. Appl Environ Microbiol 64:4180–4184. https://doi.org/10.1128/aem.64.11.4180-4184.1998
Akcil A, Koldas S (2006) Acid mine drainage (AMD): causes, treatment and case studies. J Clean Prod 14:1139–1145. https://doi.org/10.1016/j.jclepro.2004.09.006
Ro WFM (2010) Handbook of hydrocarbon and lipid microbiology. Handb Hydrocarb Lipid Microbiol. https://doi.org/10.1007/978-3-540-77587-4
Oie CSI, Albaugh CE, Peyton BM (2007) Benzoate and salicylate degradation by Halomonas campisalis, an alkaliphilic and moderately halophilic microorganism. Water Res 41:1235–1242. https://doi.org/10.1016/j.watres.2006.12.029
Xie N, Tang H, Feng J et al (2009) Characterization of benzoate degradation by newly isolated bacterium Pseudomonas sp. XP-M2. Biochem Eng J 46:79–82. https://doi.org/10.1016/j.bej.2009.04.019
Zhong W, Zhu C, Shu M et al (2010) Degradation of nicotine in tobacco waste extract by newly isolated Pseudomonas sp. ZUTSKD. Bioresour Technol 101:6935–6941. https://doi.org/10.1016/j.biortech.2010.03.142
Li J, Wang J, Li S et al (2019) Co-occurrence of functional modules derived from nicotine-degrading gene clusters confers additive effects in Pseudomonas sp. JY-Q. Appl Microbiol Biotechnol 103:4499–4510. https://doi.org/10.1007/s00253-019-09800-4
Wang SN, Xu P, Tang HZ et al (2004) Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol Lett 26:1493–1496. https://doi.org/10.1023/B:BILE.0000044450.16235.65
Wang SN, Liu Z, Tang HZ et al (2007) Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153:1556–1565. https://doi.org/10.1099/mic.0.2006/005223-0
Tang H, Wang L, Wang W et al (2013) Systematic unraveling of the unsolved pathway of nicotine degradation in pseudomonas. PLoS Genet. https://doi.org/10.1371/journal.pgen.1003923
Hu H, Wang L, Wang W et al (2019) Regulatory mechanism of nicotine degradation in pseudomonas putida. MBio. https://doi.org/10.1128/MBIO.00602-19
Yu H, Tang H, Wang L et al (2011) Complete genome sequence of the nicotine-degrading Pseudomonas putida strain S16. J Bacteriol 193:5541–5542. https://doi.org/10.1128/JB.05663-11
Ke Q, Zhang Y, Wu X et al (2018) Sustainable biodegradation of phenol by immobilized Bacillus sp. SAS19 with porous carbonaceous gels as carriers. J Environ Manag 222:185–189. https://doi.org/10.1016/j.jenvman.2018.05.061
Busca G, Berardinelli S, Resini C, Arrighi L (2008) Technologies for the removal of phenol from fluid streams: a short review of recent developments. J Hazard Mater 160:265–288. https://doi.org/10.1016/j.jhazmat.2008.03.045
Christen P, Vega A, Casalot L et al (2012) Kinetics of aerobic phenol biodegradation by the acidophilic and hyperthermophilic archaeon Sulfolobus solfataricus 98/2. Biochem Eng J 62:56–61. https://doi.org/10.1016/j.bej.2011.12.012
Agarry SE, Durojaiye AO, Solomon BO (2008) Microbial degradation of phenols: a review. Int J Environ Pollut 32:12–28. https://doi.org/10.1504/IJEP.2008.016895
Jiang Y, Yang K, Wang H et al (2015) Characteristics of phenol degradation in saline conditions of a halophilic strain JS3 isolated from industrial activated sludge. Mar Pollut Bull 99:230–234. https://doi.org/10.1016/j.marpolbul.2015.07.021
Jiang Y, Shang Y, Yang K, Wang H (2016) Phenol degradation by halophilic fungal isolate JS4 and evaluation of its tolerance of heavy metals. Appl Microbiol Biotechnol 100:1883–1890. https://doi.org/10.1007/s00253-015-7180-2
Méndez-García C, Peláez AI, Mesa V et al (2015) Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:1–17. https://doi.org/10.3389/fmicb.2015.00475
Christen P, Davidson S, Combet-Blanc Y, Auria R (2011) Phenol biodegradation by the thermoacidophilic archaeon Sulfolobus solfataricus 98/2 in a fed-batch bioreactor. Biodegradation 22:475–484. https://doi.org/10.1007/s10532-010-9420-6
Izzo V, Notomista E, Picardi A et al (2005) The thermophilic archaeon Sulfolobus solfataricus is able to grow on phenol. Res Microbiol 156:677–689. https://doi.org/10.1016/j.resmic.2005.04.001
Cox HHJ, Moerman RE, Van Baalen S et al (1997) Performance of a styrene-degrading biofilter containing the yeast Exophiala jeanselmei. Biotechnol Bioeng 53:259–266. https://doi.org/10.1002/(SICI)1097-0290(19970205)53:3%3c259::AID-BIT3%3e3.0.CO;2-H
Prenafeta-Boldú FX, Kuhn A, Luykx DMAM et al (2001) Isolation and characterisation of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycol Res 105:477–484. https://doi.org/10.1017/S0953756201003719
Qi B, Moe W, Kinney K (2002) Biodegradation of volatile organic compounds by five fungal species. Appl Microbiol Biotechnol 58:684–689. https://doi.org/10.1007/s00253-002-0938-3
Estévez E, Veiga MC, Kennes C (2005) Biodegradation of toluene by the new fungal isolates Paecilomyces variotii and Exophiala oligosperma. J Ind Microbiol Biotechnol 32:33–37. https://doi.org/10.1007/s10295-004-0203-0
Dore SY, Clancy QE, Rylee SM, Kulpa CF (2003) Naphthalene-utilizing and mercury-resistant bacteria isolated from an acidic environment. Appl Microbiol Biotechnol 63:194–199. https://doi.org/10.1007/s00253-003-1378-4
Uyttebroek M, Vermeir S, Wattiau P et al (2007) Characterization of cultures enriched from acidic polycyclic aromatic hydrocarbon-contaminated soil for growth on pyrene at low pH. Appl Environ Microbiol 73:3159–3164. https://doi.org/10.1128/AEM.02837-06
Acknowledgements
The authors would like to acknowledge the Curtin Malaysia Postgraduate Research scheme (CMPRS) under the Curtin University Malaysia for providing research fund to conduct this research.
Funding
The authors have no relevant financial or non-financial interest to disclose.
Author information
Authors and Affiliations
Contributions
SR: writing original draft and revision; TH: conceptualization, supervision, revision; SYL: review, supervision, editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Razia, S., Hadibarata, T. & Lau, S.Y. Acidophilic microorganisms in remediation of contaminants present in extremely acidic conditions. Bioprocess Biosyst Eng 46, 341–358 (2023). https://doi.org/10.1007/s00449-022-02844-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02844-3