Abstract
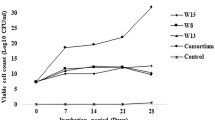
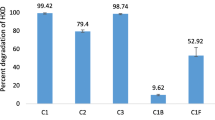
A major hindrance to the effective use of fungi in bioremediation is their inherent slow growth. Despite this, Aspergillus spp. may be used effectively. Our experiments demonstrate that bacteria, although inefficient in hydrocarbon degradation, may be effectively used in a consortium to overcome the lag in fungal utilization of petroleum hydrocarbons. Crude petroleum oil (160 mg; at 8 g/L) in minimal medium was inoculated with a previously isolated biofilm-forming consortium (Aspergillus sp. MM1 and Bacillus sp. MM1) as well as monocultures of each organism and incubated at 30 ℃ under static conditions. Residual oil was analyzed by GC–MS. Crude oil utilization of Aspergillus–Bacillus biofilm was 24 ± 1.4% in 3 days, increased to 66 ± 7% by day 5 and reached 99 ± 0.2% in 7 days. Aspergillus sp. MM1 monoculture degraded only 14 ± 6% in 5 days. However, at the end of 7 days, it was able to utilize 98 ± 2%. Bacillus sp. MM1 monoculture utilized 20 ± 4% in 7 days. This study indicates that there is a reduction of the fungal lag in bioremediation when it is in association with the bacterium. Although in monoculture, Bacillus sp. MM1 is inefficient in crude oil degradation, it synergistically enhances the initial rate of crude petroleum oil degradation of the fungus in the consortium. The rapid initial removal of as much crude oil as possible from contaminated sites is vital to minimize detrimental impacts on biodiversity.








Similar content being viewed by others
Availability of data and material
The chromatograms supporting the conclusions of this article are included as ESM1–4. Other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ezeji UE, Anyadoh SO, Ibekwe VI (2007) Clean up of crude oil-contaminated soil. Terr Aquat Environ Toxicol 1:54–59
Tissot BP, Welte DH (1978) The composition and classification of crude oils and the influence of geological factors. Petroleum formation and occurrence: a new approach to oil and gas exploration, 2nd edn. Springer, Berlin, pp 333–377
Macías-Zamora JV (2011) Ocean pollution. In: Letcher TM, Vallero DA (eds) Waste, 1st edn. Academic Press, London, pp 265–279
Hegazi AH, El-Gayar MS (2017) Role of non-hydrocarbon constituents in crude oils correlation and heavy fractions processing studies. Pet Chem 57:838–842. https://doi.org/10.1134/S0965544117100103
Freedman B (2018) Oil spills. In: Environmental science: a Canadian perspective. Dalhousie University Libraries Digital Editions
Brusseau ML (2019) Soil and groundwater remediation. In: Brusseau ML, Pepper IL, Charles PG (eds) Environmental and pollution science, 3rd edn. Elsevier, New York, pp 329–354
Watson JG (1996) Physical/chemical treatment of organically contaminated soils and sediments. J Air Waste Manag Assoc 46:993–1003. https://doi.org/10.1080/10473289.1996.10467536
Jain PK, Gupta VK, Gaur RK, Lowry M, Jaroli DPCU (2011) Bioremediation of petroleum oil contaminated soil and water. Res J Environ Toxicol 5:1–26. https://doi.org/10.3923/rjet.2011.1.26
Fondi M, Maida I, Perrin E et al (2016) Genomic and phenotypic characterization of the species Acinetobacter venetianus. Sci Rep 6:21985–21997. https://doi.org/10.1038/srep21985
Lee DW, Lee H, Kwon BO et al (2018) Biosurfactant-assisted bioremediation of crude oil by indigenous bacteria isolated from Taean beach sediment. Environ Pollut 241:254–264. https://doi.org/10.1016/j.envpol.2018.05.070
Elumalai P, Parthipan P, Karthikeyan OP, Rajasekar A (2017) Enzyme-mediated biodegradation of long-chain n -alkanes (C32 and C40) by thermophilic bacteria. 3 Biotech 7:116–126. https://doi.org/https://doi.org/10.1007/s13205-017-0773-y
Chen Q, Li J, Liu M et al (2017) Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS ONE 12:e0174445. https://doi.org/10.1371/journal.pone.0174445
Wang D, Lin J, Lin J et al (2019) Biodegradation of petroleum hydrocarbons by Bacillus subtilis BL-27, a strain with weak hydrophobicity. Molecules 24:3021–3036. https://doi.org/10.3390/molecules24173021
Sun S, Li S, Avera BN et al (2017) Soil bacterial and fungal communities show distinct recovery patterns during forest ecosystem restoration. Front Microbiol 83:966. https://doi.org/10.1128/AEM.00966-17
Wemedo SA, Nrior RR, Ike AA (2018) Biodegradation potential of Aspergillus Niger and Rhizopus arrhizus isolated from crude oil spilled site in Rivers State. J Environ Sci Toxicol Food Technol 12:49–57. https://doi.org/10.9790/2402-1212014957
AI-Jawhari IFH (2014) Ability of Some Soil Fungi in Biodegradation of Petroleum Hydrocarbon. J Appl Environ Microbiol 2:46–52. https://doi.org/10.12691/jaem-2-2-3
Al-Hawash AB, Zhang X, Ma F (2019) Removal and biodegradation of different petroleum hydrocarbons using the filamentous fungus Aspergillus sp RFC-1. Microbiologyopen 8:e619. https://doi.org/10.1002/mbo3.619
Kato T, Haruki M, Imanaka T et al (2001) Isolation and characterization of long-chain-alkane degrading Bacillus thermoleovorans from deep subterranean petroleum reservoirs. J Biosci Bioeng 91:64–70. https://doi.org/10.1016/S1389-1723(01)80113-4
Maamar A, Lucchesi ME, Debaets S et al (2020) Highlighting the crude oil bioremediation potential of marine fungi isolated from the Port of Oran (Algeria). Diversity 12:196–215. https://doi.org/10.3390/D12050196
Singh R, Paul D, Jain RK (2006) Biofilms: implications in bioremediation. Trends Microbiol 14:389–397. https://doi.org/10.1016/j.tim.2006.07.001
Davey ME, O’toole GA, (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. https://doi.org/10.1128/MMBR.64.4.847-867.2000
Edwards SJ, Kjellerup BV (2013) Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl Microbiol Biotechnol 97:9909–9921. https://doi.org/10.1007/s00253-013-5216-z
Seneviratne G, Zavahir JS, Bandara WMMS, Weerasekara MLMAW (2008) Fungal-bacterial biofilms: their development for novel biotechnological applications. World J Microbiol Biotechnol 24:739–743. https://doi.org/10.1007/s11274-007-9539-8
Benoit I, van den Esker MH, Patyshakuliyeva A et al (2015) Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ Microbiol 17:2099–2113. https://doi.org/10.1111/1462-2920.12564
Perera M, Wijayarathna D, Wijesundera S et al (2019) Biofilm mediated synergistic degradation of hexadecane by a naturally formed community comprising Aspergillus flavus complex and Bacillus cereus group. BMC Microbiol 19:1–9. https://doi.org/10.1186/s12866-019-1460-4
Bushnell LD, Haas HF (1940) The utilization of certain hydrocarbons by microorganisms. J Bacteriol 41:653–673
Dasgupta D, Ghosh R, Sengupta TK (2013) Biofilm-mediated enhanced crude oil degradation by newly isolated Pseudomonas species. Int Sch Res Not 2013:e250749. https://doi.org/10.5402/2013/250749
Barnes NM, Khodse VB, Lotlikar NP et al (2018) Bioremediation potential of hydrocarbon-utilizing fungi from select marine niches of India. 3 Biotech 8:1–10. https://doi.org/10.1007/s13205-017-1043-8
Fingas MF (1997) Studies on the evaporation of crude oil and petroleum products. II Boundary layer regulation. J Hazard Mater 57:41–58. https://doi.org/10.1016/S0304-3894(97)00051-4
WHO (1989) IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans; occupational exposures in petroleum refining; crude oil and major petroleum fuels. IARC Press, Lyon
Jeffrey AWA, McLoughlin PW, Pirkle RJ (2016) Application of isotopic compositions in fugitive petroleum product identification and correlation. Stand Handb Oil Spill Environ Forensics Fingerprinting Source Identif Second Ed. https://doi.org/10.1016/B978-0-12-809659-8.00010-3
Atlas RM (1975) Effects of temperature and crude oil composition petroleum biodegradation. Appl Microbiol 30:396–403
Brewster CS, Sharma VK, Cizmas L, McDonald TJ (2018) Occurrence, distribution and composition of aliphatic and polycyclic aromatic hydrocarbons in sediment cores from the Lower Fox River, Wisconsin, US. Environ Sci Pollut Res 25:4974–4988. https://doi.org/10.1007/s11356-017-0819-z
Neff JM, Durell GS (2012) Bioaccumulation of petroleum hydrocarbons in arctic amphipods in the oil development area of the Alaskan beaufort sea. Integr Environ Assess Manag 8:301–319. https://doi.org/10.1002/ieam.1247
Ekblad A, Wallander H, Näsholm T (1998) Chitin and ergosterol combined to measure total and living fungal biomass in ectomycorrhizas. New Phytol 138:143–149. https://doi.org/10.1046/j.1469-8137.1998.00891.x
Acknowledgements
We thank Ms. K. P. K. R. I. Dilani, Instrument Center—Faculty of Applied Sciences (IC-FAS), University of Sri Jayewardenepura (USJP), Nugegoda, Sri Lanka for technical assistance with GC–MS analysis
Funding
This work was funded by the research grant AP/3/2/2014/RG/12, University of Colombo.
Author information
Authors and Affiliations
Contributions
SJ and SW conceptualized and designed the study. MP contributed to the study design, carried out all the experimental work, interpreted the data and wrote the manuscript. SDMC supervised the GC–MS analysis. DW, GS contributed to data interpretation. All authors contributed to manuscript editing and approved the final version.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Perera, M., Chinthaka, S.D.M., Wijayarathna, C.D. et al. Reduction of lag in crude oil degradation by Aspergillus when it is in synergy with Bacillus in biofilm mode. Bioprocess Biosyst Eng 44, 1501–1510 (2021). https://doi.org/10.1007/s00449-021-02534-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-021-02534-6