Abstract
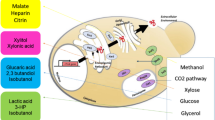
Chinese hamster ovary (CHO) cells are the main workhorse in the biopharmaceutical industry for the production of recombinant proteins, such as monoclonal antibodies. To date, a variety of metabolic engineering approaches have been used to improve the productivity of CHO cells. While genetic manipulations are potentially laborious in mammalian cells, rational design of CHO cell culture medium or efficient fed-batch strategies are more popular approaches for bioprocess optimization. In this study, a genome-scale metabolic network model of CHO cells was used to design feeding strategies for CHO cells to improve monoclonal antibody (mAb) production. A number of metabolites, including threonine and arachidonate, were suggested by the model to be added into cell culture medium. The designed composition has been experimentally validated, and then optimized, using design of experiment methods. About a two-fold increase in the total mAb expression has been observed using this strategy. Our approach can be used in similar bioprocess optimization problems, to suggest new ways of increasing production in different cell factories.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The metabolic model of CHO cells is publicly available in the supporting information of the original article (Hefzi et al. Cell Systems, 2016), which has been cited in our article. The cell line which has been used in our study is available in Radin Biotech Company of Iran.
Abbreviations
- CHO:
-
Chinese hamster ovary
- mAbs:
-
Monoclonal antibodies
- GEMs:
-
Genome-scale metabolic network models
- GPR:
-
Gene–protein–reaction
- FVSEOF:
-
Flux variability scanning based on enforced objective flux
- DoE:
-
Design of experiment
- PB:
-
Plackett–Burman
- RSM:
-
Response surface methodology
- CCD:
-
Central composite design
References
Walsh G (2014) Biopharmaceutical benchmarks 2014. Nat Biotechnol 32(10):992
Weiner GJ (2015) Building better monoclonal antibody-based therapeutics. Nat Rev Cancer 15(6):361–370
Lim Y, Wong NS, Lee YY, Ku SC, Wong DC, Yap MG (2010) Engineering mammalian cells in bioprocessing–current achievements and future perspectives. Biotechnol Appl Biochem 55(4):175–189
Kim JY, Kim Y-G, Lee GM (2012) CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol 93(3):917–930
Richelle A, Lewis NE (2017) Improvements in protein production in mammalian cells from targeted metabolic engineering. Current Opinion in Systems Biology 6:1–6
Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ (2013) The emerging CHO systems biology era: harnessing the ‘omics revolution for biotechnology. Curr Opin Biotechnol 24(6):1102–1107
Farrell A, McLoughlin N, Milne JJ, Marison IW, Bones J (2014) Application of multi-omics techniques for bioprocess design and optimization in Chinese hamster ovary cells. J Proteome Res 13(7):3144–3159
Gao Y, Ray S, Dai S, Ivanov AR, Abu-Absi NR, Lewis AM, Huang Z, Xing Z, Borys MC, Li ZJ (2016) Combined metabolomics and proteomics reveals hypoxia as a cause of lower productivity on scale-up to a 5000-liter CHO bioprocess. Biotechnol J 11(9):1190–1200
Fischer S, Handrick R, Otte K (2015) The art of CHO cell engineering: a comprehensive retrospect and future perspectives. Biotechnol Adv 33(8):1878–1896
Baik JY, Dahodwala H, Oduah E, Talman L, Gemmill TR, Gasimli L, Datta P, Yang B, Li G, Zhang F (2015) Optimization of bioprocess conditions improves production of a CHO cell-derived, bioengineered heparin. Biotechnol J 10(7):1067–1081
Zhang H, Wang H, Liu M, Zhang T, Zhang J, Wang X, Xiang W (2013) Rational development of a serum-free medium and fed-batch process for a GS-CHO cell line expressing recombinant antibody. Cytotechnology 65(3):363–378
Mellahi K, Brochu D, Gilbert M, Perrier M, Ansorge S, Durocher Y, Henry O (2019) Process intensification for the production of rituximab by an inducible CHO cell line. Bioprocess Biosyst Eng 42(5):711–725
Pereira S, Kildegaard HF, Andersen MR (2018) Impact of CHO metabolism on cell growth and protein production: an overview of toxic and inhibiting metabolites and nutrients. Biotechnol J 13(3):1700499
Bordbar A, Monk JM, King ZA, Palsson BØ (2014) Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet 15(2):107–120
Gu C, Kim GB, Kim WJ, Kim HU, Lee SY (2019) Current status and applications of genome-scale metabolic models. Genome Biol 20(1):121
Hefzi H, Ang KS, Hanscho M, Bordbar A, Ruckerbauer D, Lakshmanan M, Orellana CA, Baycin-Hizal D, Huang Y, Ley D (2016) A consensus genome-scale reconstruction of Chinese hamster ovary cell metabolism. Cell Syst 3(5):434–443
Long MR, Ong WK, Reed JL (2015) Computational methods in metabolic engineering for strain design. Curr Opin Biotechnol 34:135–141
Park HM, Kim HU, Park JM, Lee SY, Kim TY, Kim WJ (2012) Flux variability scanning based on enforced objective flux for identifying gene amplification targets. BMC Syst Biol 6(1):106
Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S (2011) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nat Protoc 6(9):1290
Nocon J, Steiger MG, Pfeffer M, Sohn SB, Kim TY, Maurer M, Rußmayer H, Pflügl S, Ask M, Haberhauer-Troyer C (2014) Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab Eng 24:129–138
Torkashvand F, Mahboudi F, Vossoughi M, Fatemi E, Moosavi Basri SM, Vaziri B (2018) Quantitative proteomic analysis of cellular responses to a designed amino acid feed in a monoclonal antibody producing Chinese hamster ovary cell line. Iran Biomed J 22(6):385–393
Barrentine LB (1999) An introduction to design of experiments: a simplified approach. ASQ Quality Press, Milwaukee
Plackett RL, Burman JP (1946) The design of optimum multifactorial experiments. Biometrika 33(4):305–325
Vanaja K, Shobha Rani R (2007) Design of experiments: concept and applications of Plackett Burman design. Clin Res Regul Aff 24(1):1–23
Guha M, Ali SZ, Bhattacharya S (2003) Screening of variables for extrusion of rice flour employing a Plackett-Burman design. J Food Eng 57(2):135–144
Cornell JA (2011) Experiments with mixtures: designs, models, and the analysis of mixture data, vol 895. John Wiley & Sons, New York
Montgomery DC (2017) Design and analysis of experiments. John wiley & sons, New York
Box GE, Wilson KB (1992) On the experimental attainment of optimum conditions. Breakthroughs in statistics. Springer, New York, pp 270–310
Vining GG, Kowalski S (2010) Statistical methods for engineers. Cengage Learning
Torkashvand F, Vaziri B, Maleknia S, Heydari A, Vossoughi M, Davami F, Mahboudi F (2015) Designed amino acid feed in improvement of production and quality targets of a therapeutic monoclonal antibody. PLoS ONE 10(10):e0140597
Rosenthal MD (1987) Fatty acid metabolism of isolated mammalian cells. Prog Lipid Res 26(2):87–124
Kelley DS, Taylor PC, Nelson GJ, Mackey BE (1998) Arachidonic acid supplementation enhances synthesis of eicosanoids without suppressing immune functions in young healthy men. Lipids 33(2):125–130
Hammarström S (1983) Leukotrienes. Annu Rev Biochem 52(1):355–377
Needleman P, Truk J, Jakschik BA, Morrison AR, Lefkowith JB (1986) Arachidonic acid metabolism. Annu Rev Biochem 55(1):69–102
Cabral M, Martín-Venegas R, Moreno JJ (2013) Role of arachidonic acid metabolites on the control of non-differentiated intestinal epithelial cell growth. Int J Biochem Cell Biol 45(8):1620–1628
Bourre J, Faivre A, Dumont O, Nouvelot A, Loudes C, Puymirat J, Tixier-Vidal A (1983) Effect of polyunsaturated fatty acids on fetal mouse brain cells in culture in a chemically defined medium. J Neurochem 41(5):1234–1242
Habbel P, Weylandt KH, Lichopoj K, Nowak J, Purschke M, Wang J-D, He C-W, Baumgart DC, Kang JX (2009) Docosahexaenoic acid suppresses arachidonic acid-induced proliferation of LS-174T human colon carcinoma cells. World J Gastroenterol 15(9):1079–1084
Chang N-W, Wu C-T, Chen D-R, Yeh C-Y, Lin C (2013) High levels of arachidonic acid and peroxisome proliferator-activated receptor-alpha in breast cancer tissues are associated with promoting cancer cell proliferation. J Nutr Biochem 24(1):274–281
Lambremont EN, Lee T-c, Blank ML, Snyder F (1978) Δ5 desaturation of fatty acids in LM cells. Biochem Biophys Res Commun 80(4):813–818
Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Assad-Garcia N, Glass JI, Covert MW (2012) A whole-cell computational model predicts phenotype from genotype. Cell 150(2):389–401
Ranganathan S, Suthers PF, Maranas CD (2010) OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput Biol 6(4):e1000744
Acknowledgements
We have to kindly thank Samira Ahmadi and Radin Biotech Company, Iran, for gifting the CHO cell line to be used in our study.
Funding
No funding available.
Author information
Authors and Affiliations
Contributions
H.F. and S.-A.M. designed the computational studies. N.E.L. was involved in computational modeling of CHO cells metabolism. H.F., B.V., F.T., and F.M. designed the lab experiments. H.F. wrote the main manuscript. N.E.L., B.V., and S.-A.M. reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Human and animal rights statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fouladiha, H., Marashi, SA., Torkashvand, F. et al. A metabolic network-based approach for developing feeding strategies for CHO cells to increase monoclonal antibody production. Bioprocess Biosyst Eng 43, 1381–1389 (2020). https://doi.org/10.1007/s00449-020-02332-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-020-02332-6