Abstract
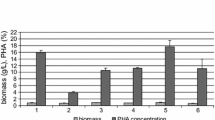
Arthrobacter sp. CGMCC 3584 is used for the industrial production of cyclic adenosine monophosphate (cAMP). However, because of the paucity of genetic engineering tools for genetic manipulation on Arthrobacter species, only a few metabolically engineered Arthrobacter have been constructed and investigated. In this study, for the first time, we constructed an arpde knockout mutant of Arthrobacter without any antibiotic resistance marker by a PCR-targeting-based homologous recombination method. Our results revealed that the deletion of arpde had little effect on biomass production and improved cAMP production by 31.1%. Furthermore, we compared the transcriptomes of the arpde knockout strain and the wild strain, aiming to understand the capacities of cAMP production due to arpde inactivation at the molecular level. Comparative transcriptomic analysis revealed that arpde inactivation had two major effects on metabolism: inhibition of glycolysis, PP pathway, and amino acid metabolism (phenylalanine, tryptophan, branched-chain amino acids, and glutamate metabolism); promotion of the purine metabolism and carbon flux from the precursor 5′-phosphoribosyl 1-pyrophosphate, which benefited cAMP production.





Similar content being viewed by others
References
Chen XC, Song H, Fang T, Cao JM, Ren HJ, Bai JX, Xiong J, Ouyang PK, Ying HJ (2010) Enhanced cyclic adenosine monophosphate production by Arthrobacter A302 through rational redistribution of metabolic flux. Biores Technol 101(9):3159–3163
Kim H, Kim S, Lee H, Park S, Lee K (2009) Expression of the cpdA gene, encoding a 3′,5′-cyclic AMP (cAMP) phosphodiesterase, is positively regulated by the cAMP–cAMP receptor protein complex. J Bacteriol 191(3):922–930
Zheng Z, Zhu M, He Y, Li N, Guo T, Chen Y, Wu J, Ying H, Xie J (2013) Gene cloning, expression, and characterization of a cyclic nucleotide phosphodiesterase from Arthrobacter sp. CGMCC 3584. Appl Biochem Biotechnol 169(8):2442–2456
Nordin K, Unell M, Jansson JK (2005) Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl Environ Microbiol 71(11):6538–6544
Niewerth H, Parschat K, Rauschenberg M, Ravoo BJ, Fetzner S (2013) The PaaX-type repressor MeqR2 of Arthrobacter sp. strain Rue61a, involved in the regulation of quinaldine catabolism, binds to its own promoter and to catabolic promoters and specifically responds to anthraniloyl coenzyme A. J Bacteriol 195(5):1068–1080
Wagenknecht M, Meinhardt F (2011) Copy number determination, expression analysis of genes potentially involved in replication, and stability assays of pAL1—the linear megaplasmid of Arthrobacter nitroguajacolicus Rü61a. Microbiol Res 166(1):14–26
Ji XJ, Huang H, Zhu JG, Ren LJ, Nie ZK, Du J, Li S (2010) Engineering Klebsiella oxytoca for efficient 2,3-butanediol production through insertional inactivation of acetaldehyde dehydrogenase gene. Appl Microbiol Biotechnol 85(6):1751–1758
Kämper J (2004) A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol Genet Genom 271(1):103–110
Ghosh S, Mohan U, Banerjee UC (2016) Studies on the production of shikimic acid using the aroK knockout strain of Bacillus megaterium. World J Microbiol Biotechnol 32(8):1–11
Chen M, Xiao X, Wang P, Zeng X, Wang F (2005) Arthrobacter ardleyensis sp. nov., isolated from Antarctic lake sediment and deep-sea sediment. Arch Microbiol 183(4):301–305
Niu H, Yang W, Zhuang K, Chen X, Chen Y, Liu D, Wu J, Zhu C, Ying H (2017) Screening of promoters from Arthrobacter sp. CGMCC 3584 using a green fluorescent protein reporter system. World J Microbiol Biotechnol 33(11):208
Sandu C, Chiribau CB, Sachelaru P, Brandsch R (2005) Plasmids for nicotine-dependent and -independent gene expression in Arthrobacter nicotinovorans and other Arthrobacter species. Appl Environ Microbiol 71(12):8920–8924
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100(4):1541–1546
Wang XL, Hou L, Zhao CG, Tang Y, Zhang B, Zhao JY, Wu YB (2019) Screening of genes involved in epithelial–mesenchymal transition and differential expression of complement-related genes induced by PAX2 in renal tubules. Nephrology 24(2):263–271
Cheng YC, Tsai RY, Sung YT, Chen IJ, Tu TY, Mao YY, Wong CS (2019) Melatonin regulation of transcription in the reversal of morphine tolerance: microarray analysis of differential gene expression. Int J Mol Med 43(2):791–806
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Shi S, Chen T, Zhang Z, Chen X, Zhao X (2009) Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab Eng 11(4):243–252
Yoshimurasuzuki T, Sagami I, Yokota N, Kurokawa H, Shimizu T (2005) DOSEc, a heme-regulated phosphodiesterase, plays an important role in the regulation of the cyclic AMP level in Escherichia coli. J Bacteriol 187(19):6678–6682
Inclan YF, Huseby MJ, Engel JN (2011) FimL regulates cAMP synthesis in Pseudomonas aeruginosa. PLoS ONE 6(1):e15867
Campoy S, Jara M, Busquets N, de Rozas AM, Badiola I, Barbé J (2002) Intracellular cyclic AMP concentration is decreased in Salmonella typhimurium fur mutants. Microbiology 148(4):1039–1048
Chen S, Segall JE (2006) EppA, a putative substrate of DdERK2, regulates cyclic AMP relay and chemotaxis in Dictyostelium discoideum. Eukaryot Cell 5(7):1136–1146
Soderling SH, Beavo JA (2000) Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr Opin Cell Biol 12(2):174–179
Matange N, Hunt DM, Buxton RS, Visweswariah SS (2013) Overexpression of the Rv0805 phosphodiesterase elicits a cAMP-independent transcriptional response. Tuberculosis 93(5):492–500
Timmermans J, Melderen LV (2010) Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 67(17):2897–2908
Banerjee A, Adolph RS, Gopalakrishnapai J, Kleinboelting S, Emmerich C, Steegborn C, Visweswariah SS (2015) A universal stress protein (USP) in mycobacteria binds cAMP. J Biol Chem 290(20):12731–12743
Miranda ER, Nam EA, Kuspa A, Shaulsky G (2015) The ABC transporter, AbcB3, mediates cAMP export in D. discoideum development. Dev Biol 397(2):203–211
Chen ZS, Lee K, Kruh GD (2001) Transport of cyclic nucleotides and estradiol 17-beta-d-glucuronide by multidrug resistance protein 4 resistance to 6-mercaptopurine and 6-thioguanine. J Biol Chem 276(36):33747–33754
Hardiman T, Lemuth K, Keller MA, Reuss M, Siemannherzberg M (2007) Topology of the global regulatory network of carbon limitation in Escherichia coli. J Biotechnol 132(4):359–374
Niu HQ, Wang JZ, Zhuang W, Liu D, Chen Y, Zhu CJ, Ying HJ (2018) Comparative transcriptomic and proteomic analysis of Arthrobacter sp. CGMCC 3584 responding to dissolved oxygen for cAMP production. Sci Rep 8(1):1246
Liu X, Yang S, Wang F, Dai X, Yang Y, Bai Z (2017) Comparative analysis of the Corynebacterium glutamicum transcriptome in response to changes in dissolved oxygen levels. J Ind Microbiol Biotechnol 44(2):181–195
Zhong Y, Xi L, Lei X, Allen HSW, Shan KH (2018) Genomic and transcriptomic comparison of Aspergillus oryzae strains: a case study in soy sauce koji fermentation. J Ind Microbiol Biotechnol 45(9):839–853
Stepansky A, Leustek T (2006) Histidine biosynthesis in plants. Amino Acids 30(2):127–142
Ingle RA (2011) Histidine biosynthesis. Arabidopsis Book 9:e0141
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(Database issue):D353–D361
Acknowledgements
This work was supported by the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (Grant No. XTE1842), the young investigator grant program of National Natural Science Foundation of China (Grant No. 21706123), the National Natural Science Foundation of China, General Program (Grant No. 31972503), the key program of the National Natural Science Foundation of China (Grant No. 21636003), Jiangsu Natural Science Fund for Distinguished Young Scholars (Grant No. BK20190035), the Program for Changjiang Scholars and Innovative Research Team in University (Grant No. IRT_14R28), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The article does not contain any researches with human participants and/or animals performed by any of the authors.
Human and animal rights
The research did not involve human participants and/or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Niu, H., Sun, X., Song, J. et al. Knockout of pde gene in Arthrobacter sp. CGMCC 3584 and transcriptomic analysis of its effects on cAMP production. Bioprocess Biosyst Eng 43, 839–850 (2020). https://doi.org/10.1007/s00449-019-02280-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02280-w