Abstract
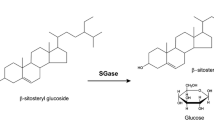
Biodiesels produced from vegetable oils have a major quality problem due to the presence of steryl glucosides (SGs), which form precipitates that clog filters and cause engine failures. Recently, we described an enzymatic process for removing SGs from biodiesel. However, industrial adoption of this technology was hindered by the cost of the steryl glucosidase (SGase) enzyme used. Here we report the development and validation at the pilot scale of a cost-efficient process for manufacturing the SGase. First, we tested various low-cost carbon sources for the Escherichia coli producing strain, ultimately developing a fed-batch fermentation process that utilizes crude glycerol as a feedstock. Next, we designed an efficient process for isolating the SGase. That process uses a novel thermolysis approach in the presence of a non-ionic detergent, centrifugation to separate the solids, and ultrafiltration to concentrate and formulate the final product. Our cost analysis indicates that on a large scale, the dose of enzyme required to eliminate SGs from each ton of biodiesel will have a manufacturing cost below $1. The new process for manufacturing the SGase, which will lead to biodiesels of a higher quality, should contribute to facilitate the global adoption of this renewable fuel. Our technology could also be used to manufacture other thermostable proteins in E. coli.






Similar content being viewed by others
References
Lane J (2014) Biofuels Mandates Around the World: 2015
OCED F (2011) OECD-FAO agricultural outlook 2011–2020. Organisation for Economic Co-operation and Development (OECD) Publishing, doi, Paris
Ringwald S (2007) Biodiesel characterization in the QC environment. The 98th AOCS annual meeting abstracts. AOCS Press, Urbana
Tang H, De Guzman R, Salley S, Ng KS (2010) Comparing process efficiency in reducing steryl glucosides in biodiesel. J Am Oil Chem Soc 87:337–345
Bondioli P, Cortesi N, Mariani C (2008) Identification and quantification of steryl glucosides in biodiesel. Eur J Lipid Sci Technol 110:120–126
Haagenson DM, Perleberg JR, Wiesenborn DP (2014) Fractionation of canola biodiesel sediment for quantification of steryl glucosides with HPLC/ELSD. J Am Oil Chem Soc 91:497–502
Pfalzgraf LLI, Foster J, Poppe G (2007) The effect of minor components on cloud point and filterability. Inf Suppl Biorenew Resour 4:17–21
Camerlynck S, Chandler J, Hornby B, van Zuylen I (2012) FAME filterability: understanding and solutions. SAE Int J Fuels Lubr 5:968–976
Plata V, Haagenson D, Dağdelen A, Wiesenborn D, Kafarov V (2015) Improvement of palm oil biodiesel filterability by adsorption methods. J Am Oil Chem Soc 92:893–903
Peiru S, Aguirre A, Eberhardt F, Braia M, Cabrera R, Menzella HG (2015) An industrial scale process for the enzymatic removal of steryl glucosides from biodiesel. Biotechnol Biofuels 8:223
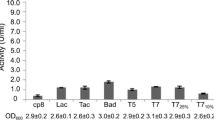
Eberhardt F, Aguirre A, Menzella HG, Peiru S (2017) Strain engineering and process optimization for enhancing the production of a thermostable steryl glucosidase in Escherichia coli. J Indus Microbiol Biotechnol 44:141–147
Jiang G, Hill DJ, Kowalczuk M, Johnston B, Adamus G, Irorere V, Radecka I (2016) Carbon sources for polyhydroxyalkanoates and an integrated biorefinery. Int J Mol Sci 17:1157
Rude MA, Schirmer A (2009) New microbial fuels: a biotech perspective. Curr Opin Microbiol 12:274–281
Ren X, Yu D, Han S, Feng Y (2007) Thermolysis of recombinant Escherichia coli for recovering a thermostable enzyme. Biochem Eng J 33:94–98
Ren X, Yu D, Yu L, Gao G, Han S, Feng Y (2007) A new study of cell disruption to release recombinant thermostable enzyme from Escherichia coli by thermolysis. J Biotechnol 129:668–673
Balasundaram B, Harrison S, Bracewell DG (2009) Advances in product release strategies and impact on bioprocess design. Trends Biotechnol 27:477–485
Koschorreck K, Wahrendorff F, Biemann S, Jesse A, Urlacher VB (2017) Cell thermolysis–A simple and fast approach for isolation of bacterial laccases with potential to decolorize industrial dyes. Process Biochem 56:171–176
Aguirre A, Peiru S, Eberhardt F, Vetcher L, Cabrera R, Menzella HG (2014) Enzymatic hydrolysis of steryl glucosides, major contaminants of vegetable oil-derived biodiesel. Appl Microbiol Biotechnol 98:4033–4040
Lee SY (1996) High cell-density culture of Escherichia coli. Trends Biotechnol 14:98–105
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences 97, 6640–6645
Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV (2004) Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proceedings of the National Academy of Sciences of the United States of America 101, 15573–15578
Yee L, Blanch H (1993) Recombinant trypsin production in high cell density fed-batch cultures in Escherichia coli. Biotechnol Bioeng 41:781–790
Eiteman MA, Altman E (2006) Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol 24:530–536
Menzella HG, Ceccarelli EA, Gramajo HC (2003) Novel Escherichia coli strain allows efficient recombinant protein production using lactose as inducer. Biotechnol Bioeng 82:809–817
Bruschi M, Krömer JO, Steen JA, Nielsen LK (2014) Production of the short peptide surfactant DAMP4 from glucose or sucrose in high cell density cultures of Escherichia coli BL21 (DE3). Microbial Cell Factories 13:99
Ramalingam S, Gautam P, Mukherjee KJ, Jayaraman G (2007) Effects of post-induction feed strategies on secretory production of recombinant streptokinase in Escherichia coli. Biochem Eng J 33:34–41
Wong HH, Kim YC, Lee SY, Chang HN (1998) Effect of post-induction nutrient feeding strategies on the production of bioadhesive protein in Escherichia coli. Biotechnol Bioeng 60:271–276
Gao H-J, Wu Q, Chen G-Q (2002) Enhanced production of D-(–)-3-hydroxybutyric acid by recombinant Escherichia coli. FEMS Microbiol Lett 213:59–65
Jensen EB, Carlsen S (1990) Production of recombinant human growth hormone in Escherichia coli: expression of different precursors and physiological effects of glucose, acetate, and salts. Biotechnol Bioeng 36:1–11
Strandberg L, Enfors S-O (1991) Batch and fed batch cultivations for the temperature induced production of a recombinant protein in Escherichia coli. Biotech Lett 13:609–614
De Anda R, Lara AR, Hernández V, Hernández-Montalvo V, Gosset G, Bolívar F, Ramírez OT (2006) Replacement of the glucose phosphotransferase transport system by galactose permease reduces acetate accumulation and improves process performance of Escherichia coli for recombinant protein production without impairment of growth rate. Metabol Eng 8:281–290
Hellmuth K, Korz D, Sanders E, Deckwer W-D (1994) Effect of growth rate on stability and gene expression of recombinant plasmids during continuous and high cell density cultivation of Escherichia coli TG1. J Biotechnol 32:289–298
Jin D, Chung BH, Do Hwang Y, Park YH (1992) Glucose-limited fed-batch culture of Escherichia coli for production of recombinant human interleukin-2 with the DO-stat method. J Ferment Bioeng 74:196–198
Junker BH (2004) Scale-up methodologies for Escherichia coli and yeast fermentation processes. J Biosci Bioeng 97:347–364
Ju L-K, Chase G (1992) Improved scale-up strategies of bioreactors. Bioprocess Biosyst Eng 8:49–53
Paul EL, Atiemo-Obeng VA, Kresta SM (2004) Handbook of industrial mixing: science and practice. John Wiley & Sons
Garcia-Ochoa F, Gomez E (2009) Bioreactor scale-up and oxygen transfer rate in microbial processes: an overview. Biotechnol Adv 27:153–176
Holmberg A, Ranta J (1982) Procedures for parameter and state estimation of microbial growth process models. Automatica 18:181–193
O’Sullivan LM, Patel S, Ward JM, Woodley JM, Doig SD (2001) Large scale production of cyclohexanone monooxygenase from Escherichia coli TOP10 pQR239. Enzyme Microb Technol 28:265–274
Xu B, Jahic M, Blomsten G, Enfors S-O (1999) Glucose overflow metabolism and mixed-acid fermentation in aerobic large-scale fed-batch processes with Escherichia coli. Appl Microbiol Biotechnol 51:564–571
Bylund F, Collet E, Enfors S-O, Larsson G (1998) Substrate gradient formation in the large-scale bioreactor lowers cell yield and increases by-product formation. Bioprocess Eng 18:171–180
Enfors S-O, Jahic M, Rozkov A, Xu B, Hecker M, Jürgen B, Krüger E, Schweder T, Hamer G, O’beirne D (2001) Physiological responses to mixing in large scale bioreactors. J Biotechnol 85:175–185
Middelberg AP, O’Neill BK, Bogle L, David I, Snoswell MA (1991) A novel technique for the measurement of disruption in high-pressure homogenization: Studies on E. coli containing recombinant inclusion bodies. Biotechnol Bioeng 38:363–370
Sauer T, Robinson CW, Glick BR (1989) Disruption of native and recombinant Escherichia coli in a high-pressure homogenizer. Biotechnol Bioeng 33:1330–1342
Matsui I, Sakai Y, Matsui E, Kikuchi H, Kawarabayasi Y, Honda K (2000) Novel substrate specificity of a membrane-bound β-glycosidase from the hyperthermophilic archaeon Pyrococcus horikoshii. FEBS letters 467:195–200
Watson J, Cumming R, Street G, Tuffnell J (1987) Release of intracellular protein by thermolysis, pp 105–109
Zhao CX, Dwyer MD, Yu AL, Wu Y, Fang S, Middelberg AP (2015) A simple and low-cost platform technology for producing pexiganan antimicrobial peptide in E. coli. Biotechnol Bioeng 112:957–964
Potvin G, Ahmad A, Zhang Z (2012) Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: a review. Biochem Eng J 64:91–105
Klein-Marcuschamer D, Simmons BA, Blanch HW (2011) Techno-economic analysis of a lignocellulosic ethanol biorefinery with ionic liquid pre-treatment. Biofuels Bioprod Biorefin 5:562–569
Janet L (2016) ENERGÍAS RENOVABLES REPORTE DE LA SITUACIÓN MUNDIAL, REN21 Paris. France
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.

Rights and permissions
About this article
Cite this article
Eberhardt, F., Aguirre, A., Paoletti, L. et al. Pilot-scale process development for low-cost production of a thermostable biodiesel refining enzyme in Escherichia coli. Bioprocess Biosyst Eng 41, 555–564 (2018). https://doi.org/10.1007/s00449-018-1890-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-018-1890-7