Abstract
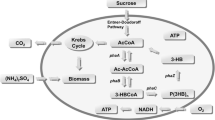
This article presents a modeling approach for industrial 2-keto-l-gulonic acid (2-KGA) fed-batch fermentation by the mixed culture of Ketogulonicigenium vulgare (K. vulgare) and Bacillus megaterium (B. megaterium). A macrokinetic model of K. vulgare is constructed based on the simplified metabolic pathways. The reaction rates obtained from the macrokinetic model are then coupled into a bioreactor model such that the relationship between substrate feeding rates and the main state variables, e.g., the concentrations of the biomass, substrate and product, is constructed. A differential evolution algorithm using the Lozi map as the random number generator is utilized to perform the model parameters identification, with the industrial data of 2-KGA fed-batch fermentation. Validation results demonstrate that the model simulations of substrate and product concentrations are well in coincidence with the measurements. Furthermore, the model simulations of biomass concentrations reflect principally the growth kinetics of the two microbes in the mixed culture.









Similar content being viewed by others
Abbreviations
- a, b :
-
Parameters of the Lozi map
- CR:
-
Crossover constant of DE
- DE:
-
Differential evolution algorithm
- D :
-
Dimension of parameter vector in DE
- e :
-
Simulation error
- f :
-
Cost function
- F O :
-
Withdrawal rate by sampling (L/h)
- F B :
-
Base solution feeding rate (L/h)
- F S :
-
Substrate feeding rate (L/h)
- F :
-
Mutation factor of DE
- GA:
-
Genetic algorithm
- m ATP :
-
Maintenance coefficient for ATP (mol/g/h)
- m 1, m 2 :
-
Mutualism coefficients
- M S :
-
Molecular weight of substrate
- NP:
-
Number of parameter vectors in a population
- N i :
-
Number of sampling points of the ith batch
- P(T k ):
-
Measurement of 2-KGA concentration at T k (g/L)
- P m(T k ):
-
Model simulation of 2-KGA concentration at T k (g/L)
- P/O :
-
Effectiveness coefficient of oxidative phosphorylation
- P :
-
Product concentration (g/L)
- r ATP :
-
Specific ATP uptake rate (mol/g/h)
- r B1, r B2 :
-
Specific bio-synthesis rate of biomass (mol/g/h)
- r G6P :
-
Specific Gluconate-6-phosphate uptake rate in glycolysis (mol/g/h)
- \(r_{{{\text{O}}_{2} }}\) :
-
Specific oxygen uptake rate (mol/g/h)
- r gr :
-
Specific Ribulose-5- phosphate production rate (mol/g/h)
- r S :
-
Specific substrate uptake rate of K. vulgare (mol/g/h)
- r S max :
-
The maximum specific substrate uptake rate in Monod model (mol/g/h)
- r S min :
-
The initial input value of r S in the regulator model (mol/g/h)
- r TCC :
-
Specific acetyl CoA uptake rate (mol/g/h)
- r AC :
-
Specific acetyl CoA production rate (mol/g/h)
- r SM :
-
Specific l-sorbose uptake rate obtained from Monod model (mol/g/h)
- r SR :
-
Specific l-sorbose uptake rate obtained from the regulator model (mol/g/h)
- r NAD :
-
Specific NADH uptake rate in the respiratory chain (mol/g/h)
- r 1, r 2 :
-
Random indexes
- RMSE:
-
Root mean square error
- S :
-
Substrate concentration in the medium (g/L)
- S R :
-
Substrate concentration in the feed (g/L)
- T k :
-
The kth sampling point of 2-KGA fermentation (h)
- u i,G+1 :
-
The ith trial vector for generation G + 1 of DE
- V :
-
Culture volume (L)
- v i,G+1 :
-
The ith mutant vector for generation G + 1 of DE
- X B :
-
B. megaterium concentration (g/L)
- X K :
-
K. vulgare concentration (g/L)
- X B max :
-
Maximum concentration of B. megaterium (g/L)
- X K max :
-
Maximum concentration of K. vulgare (g/L)
- X n+1 :
-
Random number generated by the Lozi map
- x i,G :
-
The ith parameter vector for generation G of DE
- x Best,G :
-
Best performed parameter vector in generation G
- Y ATP :
-
Yield coefficient of ATP (g/mol)
- Y n+1 :
-
Random number generated by the Lozi map
- Y P/S :
-
Product yield coefficient (g/g)
- μ 1 :
-
Specific growth rate of B. megaterium (/h)
- μ 2 :
-
Specific growth rate of K. vulgare (/h)
- μ b1 :
-
Specific B. megaterium substance uptake rate by K. vulgare (mol/g/h)
- γ :
-
Coefficient of evaporation (/h)
- K 21, K 22, α, β, K B1, K B2, K S2 :
-
Model parameters
References
Reichstein T, Grüssner A (1934) Eine ergiebige Synthese der l-Ascorbinsäure (C-Vitamin). Helv Chim Acta 17:311–328
Ji AG, Gao PJ (1998) Synthesis of 2-keto-l-gulonic acid from gluconic acid by co-immobilized Gluconobacter oxydans and Corynebacterium sp. Biotechnol Lett 20:939–942
Urbance JW, Bratina BJ, Stoddard SF (2001) Taxonomic characterization of Ketogulonigenium vulgare gen. nov., sp. nov. and Ketogulonigenium robustum sp. nov., which oxidize L-sorbose to 2-keto-l-gulonic acid. Int J Syst Evol Micr 51:1059–1070
Zhang J, Liu J, Shi ZP, Liu LM, Chen J (2010) Manipulation of B-megaterium growth for efficient 2-KLG production by K-vulgare. Process Biochem 45:602–606
Lee HW, Pan JG (1999) Screening for L-sorbose and L-sorbosone dehydrogenase producing microbes for 2-keto-l-gulonic acid production. J Ind Microb Biotech 23:106–111
Liu LM, Chen KJ, Zhang J, Liu J, Chen J (2011) Gelatin enhances 2-keto-l-gulonic acid production based on Ketogulonigenium vulgare genome annotation. J Biotechnol 156:182–187
Zhou J, Ma Q, Yi H, Wang L, Song H, Yuan YJ (2011) Metabolome profiling reveals metabolic cooperation between Bacillus megaterium and Ketogulonicigenium vulgare during induced swarm motility. Appl Environ Microb 77:7023–7030
Dietz D, Zeng AP (2014) Efficient production of 1, 3-propanediol from fermentation of crude glycerol with mixed cultures in a simple medium. Bioproc Biosyst Eng 37:225–233
Dietz D, Sabra W, Zeng AP (2013) Co-cultivation of Lactobacillus zeae and Veillonella criceti for the production of propionic acid. AMB Express 3:1–9
Wei DZ, Yuan WK, Yin GL (1992) Studies on kinetic model of vitamin C two-step fermentation process. Chin J Biotechnol 8:277–282
Zhang ZX, Zhu XJ, Xie P, Sun JW, Yuan JQ (2012) Macrokinetic model for Gluconobacter oxydans in 2-keto-l-gulonic acid mixed culture. Biotechnol Bioeng 17:1008–1017
Yang F, Jia Q, Xiong ZH, Zhang XB, Wu HT, Zhao Y, Yang J, Zhu JP, Dong J, Xue Y, Sun LL, Shen Y, Jin Q (2006) Complete genome analysis of Ketogulonigenium sp.WB0104. Chinese Sci Bull 51:941–945
Xu A, Yao JM, Yu ZL (1998) Improvement of mingle strains in the fermentation of 2-keto-l-gulonic acid—precursor of Vc by ion implantation. Ind Microbiol 28:21–24
Nakamura M (1968) Determination of fructose in the presence of a large excess of glucose, part V. A modified cysteine-carbazole reaction. Agric Biol Chem 32:701–706
Hoshino T, Sugisawa T, Tazoe M, Shinjoh M, Fujiwara A (1990) Metabolic pathway for 2-Keto-l-gulonic acid formation in gluconobacter melanogenus IFO3293. Agri Biol Chem 54:1211–1218
Sonnleitner B, Kaeppeli O (1986) Growth of Saccharomyces cerevisiae is controlled by its limited respiratory capacity: formulation and verification of a hypothesis. Biotechnol Bioeng 28:927–937
Yuan JQ, Liu YH, Geng J (2010) Stoichiometric balance based macrokinetic model for Penicillium chrysogenum in fed-batch fermentation. Process Biochem 45:542–548
Luedeking R, Piret EL (2000) A kinetic study of the lactic acid fermentation. Batch process at controlled pH. Biotechnol Bioeng 67:636–644
Ren HT, Yuan JQ, Bellgardt KH (2003) Macrokinetic model for methylotrophic Pichia pastoris based on stoichiometric balance. J Biotechnol 106:53–68
Cosenza A, Mannina G, Neumann MB, Viviani G, Vanrolleghem PA (2013) Biological nitrogen and phosphorus removal in membrane bioreactors: model development and parameter estimation. Bioproc Biosyst Eng 36:499–514
Votruba J (1982) Practical aspects of the mathematical modeling of fermentation processes as a method of description, simulation, identification and optimization. Acta Biotechnol 2:119–126
Andrade RR, Rivera EC, Atala IPD, Filho RM, Filho FM, Costa AC (2009) Study of kinetic parameters in a mechanistic model for bioethanol production through a screening technique and optimization. Bioproc Biosyst Eng 32:673–680
Rainer S, Kenneth P (1997) Differential evolution—a simple and efficient heuristic for global optimization over continuous spaces. J Global Optim 11:341–359
Qin AK, Huang VL, Suganthan PN (2009) Differential evolution algorithm with strategy adaptation for global numerical optimization. Evol Comput 13:398–417
Das S, Suganthan PN (2011) Differential evolution: a survey of the state-of-the-art. Evol Comput 15:4–31
Caponetto R, Fortuna L, Fazzino S, Xibilia MG (2003) Chaotic sequences to improve the performance of evolutionary algorithms. Evol Comput 7:289–304
Pluhacek M, Senkerik R, Davendra D, Oplatkova ZK, Zelinka I (2012) On the behavior and performance of chaos driven PSO algorithm with inertia weight. Comput Math Appl 66:122–134
Nickabadi A, Ebadzadeh MM, Safabakhsh R (2011) A novel particle swarm optimization algorithm with adaptive inertia weight. Appl Soft Comput 11:3658–3670
Roeva O (2006) A modified genetic algorithm for a parameter identification of fermentation processes. Biotechnol Biotec Eq 20:202–209
Acknowledgments
This study is supported by the Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology/Chinese Academy of Sciences,the Doctoral Program of Higher Education of China (Grant No. 20110073110018) and the National Science Foundation of China (Grant No. 61233004). J.Q.Yuan acknowledges the financial support of Alexander von Humboldt-Stiftung/Germany during the early stage of the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, T., Sun, J. & Yuan, J. Modeling and parameters identification of 2-keto-l-gulonic acid fed-batch fermentation. Bioprocess Biosyst Eng 38, 605–614 (2015). https://doi.org/10.1007/s00449-014-1300-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-014-1300-8