Abstract
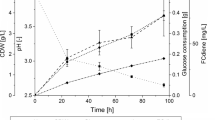
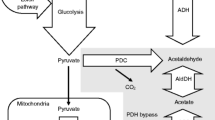
Saccharomyces cerevisiae CEN.PK113-5D, a strain auxotrophic for uracil belonging to the CEN.PK family of the yeast S. cerevisiae, was cultured in aerated fed-batch reactor as such and once transformed to express human interleukin-1β (IL-1β), aiming at obtaining high cell densities and optimizing IL-1β production. Three different exponentially increasing glucose feeding profiles were tested, all of them “in theory” promoting respiratory metabolism to obtain high biomass/product yield. A non-structured non-segregated model was developed to describe the performance of S. cerevisiae CEN.PK113-5D during the fed-batch process and, in particular, its capability to metabolize simultaneously glucose and ethanol which derived from the precedent batch growth. Our study showed that the proliferative capacity of the yeast population declined along the fed-batch run, as shown by the exponentially decreasing specific growth rates on glucose. Further, a shift towards fermentative metabolism occurred. This shift took place earlier the higher was the feed rate and was more pronounced in the case of the recombinant strain. Determination of some physiological markers (acetate production, intracellular ROS accumulation, catalase activity and cell viability) showed that neither poor oxygenation nor oxidative stress was responsible for the decreased specific growth rate, nor for the shift to fermentative metabolism.




Similar content being viewed by others
Abbreviations
- \(\beta\) :
-
Inhibition factor of glucose on ethanol consumption
- \(E\) :
-
Ethanol concentration at time \(t\) (g L−1)
- \(E_{0}\) :
-
Ethanol concentration at the end of batch phase/beginning of fed-batch phase (g L−1)
- \(F\) :
-
Glucose flow rate at time \(t\) (L h−1)
- \(F_{0}\) :
-
Glucose flow rate at the beginning of fed-batch phase (L h−1)
- \(G\) :
-
Residual glucose concentration at time \(t\) (g L−1)
- \(G_{0}\) :
-
Residual glucose concentration at the beginning of fed-batch phase (g L−1)
- \(G_{\text{R}}\) :
-
Glucose concentration in the feeding (g L−1)
- \(k_{\text{d}}\) :
-
Specific death rate (h−1)
- \(\left( {k_{S} } \right)_{E}\) :
-
Ethanol saturation constant (g L−1)
- \(\left( {\mu_{ \hbox{max} } } \right)_{E}\) :
-
Maximum specific growth rate on ethanol (h−1)
- \(\left( {\mu_{G} } \right)_{i}\) :
-
Specific growth rate on glucose in the i th interval (h−1)
- \(t\) :
-
Arbitrary time of fed-batch run (h−1)
- \(t_{\text{EC}}\) :
-
Time of fed-batch run when ethanol finished to be consumed and the variation of specific growth rate on glucose equation occurs (h)
- \(t_{\text{EP}}\) :
-
Time of fed-batch run when ethanol begins to be accumulated (h)
- \(V\) :
-
Broth culture volume (L)
- \(X\) :
-
Viable biomass concentration at time \(t\) (g cell d.w. L−1)
- \(X_{0}\) :
-
Initial viable biomass concentration (g cell d.w. L−1)
- \(\left( {Y_{X/G} } \right)_{i}\) :
-
Biomass yield on glucose in the i-th interval of fed-batch run (g cell d.w. g glucose−1)
- \(\left( {Y_{X/G} } \right)_{1}\) :
-
Biomass yield on glucose in the interval 0 ≤ t < t EC (g cell d.w. g glucose−1)
- \(\left( {Y_{X/G} } \right)_{2}\) :
-
Biomass yield on glucose in the interval t ≥ t EC (g cell d.w. g glucose−1)
- \(\left( {Y_{X/E} } \right)_{S}\) :
-
Biomass yield on ethanol as substrate in the interval 0 ≤ t < t EC (g cell d.w. g ethanol−1)
- \(\left( {Y_{E/X} } \right)_{P}\) :
-
Ethanol produced per unit of biomass in the interval t ≥ t EP (g ethanol g cell d.w.−1)
References
Riesenberg D, Guthke R (1999) High-cell-density cultivation of microorganisms. Appl Microbiol Biotechnol 51(4):422–430
Shojaosadati SA, Varedi SMK, Babaei Pour V, Farnoud AM (2008) Recent advances in high cell density cultivation for production of recombinant protein. Iran J Biotechnol 6:63–84
Martinez JL, Liu L, Petranovic D, Nielsen J (2012) Pharmaceutical protein production by yeast: towards production of human blood proteins by microbial fermentation. Curr Opin Biotechnol 23:965–971
Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D (2012) Recombinant protein production in yeasts. Methods Mol Biol 824:329–358
Beudeker RF, van Dam HW, van der Plaat JB, Vellega K (1990) Developments in bakers’ yeast production. In: Verachtert H, De Mot R (eds) Yeast biotechnology and biocatalysis. Marcel Dekker, New York
Landi C, Paciello L, de Alteriis E, Brambilla L, Parascandola P (2011) Effect of auxotrophies on yeast performance in aerated fed-batch reactor. Biochem Biophys Res Comm 414:604–611
Andrés I, Gallardo O, Parascandola P, Pastor J, Zueco J (2005) Use of the cell wall protein Pir4 as a fusion partner for the expression of Bacillus sp.BP-7 xylanase A in Saccharomyces cerevisiae. Biotechnol Bioeng 89:690–697
Moukadiri I, Jafar L, Zueco J (1999) Identification of two mannoproteins released from cell walls of Saccharomyces cerevisiae mnn1 and mnn9 double mutant by reducing agents. J Bacteriol 181:4741–4745
Paciello L, Andrès I, Zueco J, Bianchi MM, de Alteriis E, Parascandola P (2010) Expression of human interleukin-1β in Saccharomyces cerevisiae using PIR4 as fusion partner and production in aerated fed-batch reactor. Ann Microbiol 60:719–728
Enfors SO, Häggström L (1998) Bioprocess technology: fundamentals and applications. Royal Institute of Technology, Stockholm
Porro D, Brambilla L, Alberghina L (2003) Glucose metabolism and cell size in continuous cultures of Saccharomyces cerevisiae. FEMS Microbiol Lett 229:165–171
Verduyn C, Postma E, Scheffers WA, van Dijken JP (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517
Pronk JT (2002) Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol 68:2095–2100
Paciello L, de Alteriis E, Mazzoni C, Palermo V, Zueco J, Parascandola P (2009) Performance of the auxotrophic Saccharomyces cerevisiae BY4741 as host for human interleukin-1β production in an aerated fed-batch reactor. Microb Cell Fact 8:70–82
Enfors SO (2001) Baker’s yeast. In: Ratledge C, Kristiansen B (eds) Basic Biotechnology. Cambridge University Press, Cambridge
Camattari A, Bianchi MM, Branduardi P, Porro D, Brambilla L (2007) Induction by hypoxia of heterologous-protein production with the KlPDC1 promoter in yeasts. Appl Env Microbiol 73:922–929
Willis C (1990) Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol 25:245–280
Valadi A, Granath K, Gustafsson L, Adler L (2004) Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J Biol Chem 279(38):39677–39685
Verduyn C, Postma E, Scheffers WA, van Dijken JP (1990) Energetics of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol 136:405–412
De Deken RH (1966) The crabtree effect: a regulatory system in yeast. J Gen Microbiol 44:149–156
Petrik M, Käppeli O, Fiechter A (1983) An expanded concept for glucose effect in the yeast Saccharomyces uvarum: involvement of short- and long-term regulation. J Gen Microbiol 129:43–49
van Hoek P, Flikweert MT, van Der Aart QJM, De Steensma HY, van Dijken JP, Pronk JT (1998) Effects of pyruvate decarboxylase overproduction on flux distribution at the pyruvate branch point in Saccharomyces cerevisiae. Appl Env Microbiol 64:2133–2140
Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WHM, Klaassen P, Paddon CJ, Platt D, Kötter P, van Ham RC, Reinders MJT, Pronk JT, de Ridder D, Daran JM (2012) De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 11:36
Glick BR (1995) Metabolic load and heterologous gene expression. Biotechnol Adv 13:247–261
Gorgens JF, van Zyl WH, Knoetze JH, Hahn-Hagerdal B (2001) The metabolic burden of the PGK1 and ADH2 promoter systems for heterologous xylanase production by Saccharomyces cerevisiae in defined medium. Biotechnol Bioeng 73:238–245
Molenaar D, van Berlo R, de Ridder D, Teusink B (2009) Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol 5:323–332
Semchyshyn HM, Abrat OB, Miedzobrodzki J, Inoue Y, Lushchak VIì (2011) Acetate but not propionate induces oxidative stress in bakers’ yeast Saccharomyces cerevisiae. Redox Rep 16:15–23
Casatta N, Porro A, Orlandi I, Brambilla L, Vai M (2012) Lack of Sir2 increases acetate consumption and decreases extracellular pro-aging factors. Biochim Biophys Acta 1833:593–601
Postma E, Verduyn C, Scheffers WA, van Dijken JP (1989) Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Env Microbiol 55:468–477
Chance B, Oshino N (1971) Kinetics and mechanisms of catalase in peroxisomes of the mitochondrial fraction. Biochem J 122:225–233
Petrova VY, Rasheva TV, Kujumdzieva AV (2002) Catalase enzyme in mitochondria of Saccharomyces cerevisiae. Electron J Biotechnol 5:29–41
Acknowledgments
This research was supported by the University of Salerno funds (FARB, 2011) to Palma Parascandola in the framework of the research project “Ottimizzazione e modellazione di sistemi HCDC (high cell-density cultivation) per la produzione di masse microbiche da lievito: parametri biologici e condizioni di processo”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Landi, C., Paciello, L., de Alteriis, E. et al. High cell density culture with S. cerevisiae CEN.PK113-5D for IL-1β production: optimization, modeling, and physiological aspects. Bioprocess Biosyst Eng 38, 251–261 (2015). https://doi.org/10.1007/s00449-014-1264-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-014-1264-8